| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,146,695 – 23,146,807 |
| Length | 112 |
| Max. P | 0.972464 |

| Location | 23,146,695 – 23,146,807 |
|---|---|
| Length | 112 |
| Sequences | 13 |
| Columns | 136 |
| Reading direction | forward |
| Mean pairwise identity | 77.64 |
| Shannon entropy | 0.46510 |
| G+C content | 0.45521 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -17.72 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
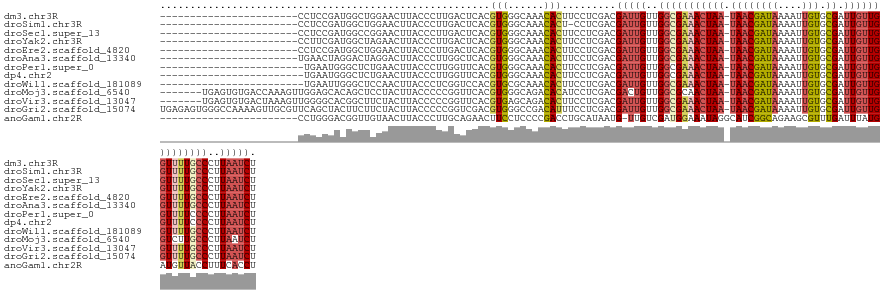
>dm3.chr3R 23146695 112 + 27905053 -----------------------CCUCCGAUGGCUGGAACUUACCCUUGACUCACGUGGGCAAACACUUCCUCGACGAUUGUUGGCGAAACUAA-UAACGAUAAAAUUGUGCGAUUGUUGGUUUUGCCCUUAAUCU -----------------------....(((.((..(.......(((.((.....)).)))......)..)))))..(((((..(((((((((((-((((((((....))).)).))))))))))))))..))))). ( -30.82, z-score = -2.89, R) >droSim1.chr3R 22895790 111 + 27517382 -----------------------CCUCCGAUGGCUGGAACUUACCCUUGACUCACGUGGGCAAACACU-CCUCGACGAUUGUUGGCGAAACUAA-UAACGAUAAAAUUGUGCGAUUGUUGGUUUUGCCCUUAAUCU -----------------------....(((.((..((.......))(((.(((....)))))).....-)))))..(((((..(((((((((((-((((((((....))).)).))))))))))))))..))))). ( -29.70, z-score = -2.55, R) >droSec1.super_13 1900081 112 + 2104621 -----------------------CCUCCGAUGGCCGGAACUUACCCUUGACUCACGUGGGCAAACACUUCCUCGACGAUUGUUGGCGAAACUAA-UAACGAUAAAAUUGUGCGAUUGUUGGUUUUGCCCUUAAUCU -----------------------..((((.....))))................((.(((........))).))..(((((..(((((((((((-((((((((....))).)).))))))))))))))..))))). ( -31.20, z-score = -2.67, R) >droYak2.chr3R 5486802 112 - 28832112 -----------------------CCUUCGAUGGCUAGAACUUACCCUUGACUCACGUGGGCAAACACUUCCUCGACGAUUGUUGGCGAAACUAA-UAACGAUAAAAUUGUGCGAUUGUUGGUUUUGCCCUUAAUCU -----------------------...((((.((..((......(((.((.....)).)))......)).)))))).(((((..(((((((((((-((((((((....))).)).))))))))))))))..))))). ( -31.80, z-score = -3.54, R) >droEre2.scaffold_4820 5585271 112 - 10470090 -----------------------CCUCCGAUGGCUGGAACUUACCCUUGACUCACGUGGGCAAACACUUCCUCGACGAUUGUUGGCGAAACUAA-UAACGAUAAAAUUGUGCGAUUGUUGGUUUUGCCCUUAAUCU -----------------------....(((.((..(.......(((.((.....)).)))......)..)))))..(((((..(((((((((((-((((((((....))).)).))))))))))))))..))))). ( -30.82, z-score = -2.89, R) >droAna3.scaffold_13340 4153269 112 - 23697760 -----------------------UGAACUAGGACUAGGACUUACCCUUGGCUCACGUGGGCAAACACUUCCUCGACGAUUGUUGGCGAAACUAA-UAACGAUAAAAUUGUGCGAUUGUUGGUUUUGCCCUUAAUCU -----------------------....(.(((((.(((......))).)((((....)))).......)))).)..(((((..(((((((((((-((((((((....))).)).))))))))))))))..))))). ( -34.10, z-score = -3.59, R) >droPer1.super_0 2608712 111 + 11822988 ------------------------UGAAUGGGCUCUGAACUUACCCUUGGUUCACGUGGGCAAACACUUCCUCGACGAUUGUUGGCGAAACUAA-UAACGAUAAAAUUGUGCGAUUGUUGGUUUUCCCCUUAAUCU ------------------------.(((((.((((((((((.......))))))...))))...)).)))......(((((..((.((((((((-((((((((....))).)).))))))))))).))..))))). ( -28.10, z-score = -1.59, R) >dp4.chr2 8796050 111 + 30794189 ------------------------UGAAUGGGCUCUGAACUUACCCUUGGUUCACGUGGGCAAACACUUCCUCGACGAUUGUUGGCGAAACUAA-UAACGAUAAAAUUGUGCGAUUGUUGGUUUUCCCCUUAAUCU ------------------------.(((((.((((((((((.......))))))...))))...)).)))......(((((..((.((((((((-((((((((....))).)).))))))))))).))..))))). ( -28.10, z-score = -1.59, R) >droWil1.scaffold_181089 3369791 111 + 12369635 ------------------------UGAAUUGGGCUCCAACUUACCCUCGGUCCACGUGCGCAAACACUUCCUCGACGAUUGUUGGCGAAACUAA-UAACGAUAAAAUUGUGCGAUUGUUGGUUUUGCCCUUAAUCU ------------------------.....((((((.............)))))).(((......))).........(((((..(((((((((((-((((((((....))).)).))))))))))))))..))))). ( -29.62, z-score = -2.15, R) >droMoj3.scaffold_6540 20120030 128 - 34148556 -------UGAGUGUGACCAAAGUUGGAGCACAGCUCCUACUUACCCCCGGUUCACGUGGGCAGACACAUCCUCGACGACUGUUGGCGCAACUAA-UAACGAUAAAAUUGUGCGAUUGUUGGUCUUGCCCUUAAUCU -------.(((((((....((((.(((((...))))).)))).(((.((.....)).)))....))))..)))...((.((..((((..(((((-((((((((....))).)).))))))))..))))..)).)). ( -36.90, z-score = -1.19, R) >droVir3.scaffold_13047 17651887 128 + 19223366 -------UGAGUGUGACUAAAGUUGGGGCACGGCUUCUACUUACCCCCGGUUCACGUGAGCAGACACUUCCUCGACGAUUGUUGGCGAAACUAA-UAACGAUAAAAUUGUGCGAUUGUUGGUUUUGCCCUUAAUCU -------.((((((.........(((((...((......))...)))))((((....))))..)))))).......(((((..(((((((((((-((((((((....))).)).))))))))))))))..))))). ( -39.10, z-score = -2.25, R) >droGri2.scaffold_15074 796448 135 - 7742996 UGAGAGUGGGCCAAAAGUUGCGUUCAGCUACUUCUUCUACUUACCCCCGGUCGACGUGGGCCGACAUUUCCUCGACGAUUGUUGGCGAAACUAA-UAACGAUAAAAUUGUGCGAUUGUUGGUUUUGCCCUUAAUCU .(((((((((....((((.((.....)).))))..))))))......(((((......))))).......)))...(((((..(((((((((((-((((((((....))).)).))))))))))))))..))))). ( -42.00, z-score = -2.39, R) >anoGam1.chr2R 7786051 112 + 62725911 -----------------------CCUGGGACGGUUGUAACUUACCCUUGCAGAACUUCCUCCCCGACCUGCAUAAUG-UUGUCGAUGGAAAUAGGCAUCGGCAGAAGCGUUUGAUUUAUGAUGUUACCUUUCACCU -----------------------...((((.((..(((((.......(((((...............))))).((((-(((((((((........))))))))...)))))...........))))))).)).)). ( -23.46, z-score = 0.75, R) >consensus _______________________CCUACGAUGGCUGGAACUUACCCUUGGCUCACGUGGGCAAACACUUCCUCGACGAUUGUUGGCGAAACUAA_UAACGAUAAAAUUGUGCGAUUGUUGGUUUUGCCCUUAAUCU .......................................................(((......))).........(((((..(((((((((((....(((((....))).))....)))))))))))..))))). (-17.72 = -18.12 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:44 2011