| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,119,083 – 23,119,177 |
| Length | 94 |
| Max. P | 0.789439 |

| Location | 23,119,083 – 23,119,177 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
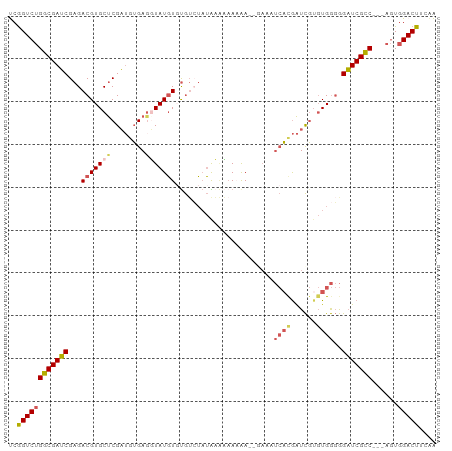
| Mean pairwise identity | 71.97 |
| Shannon entropy | 0.49232 |
| G+C content | 0.49166 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -17.30 |
| Energy contribution | -18.14 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.789439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
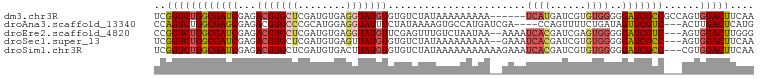
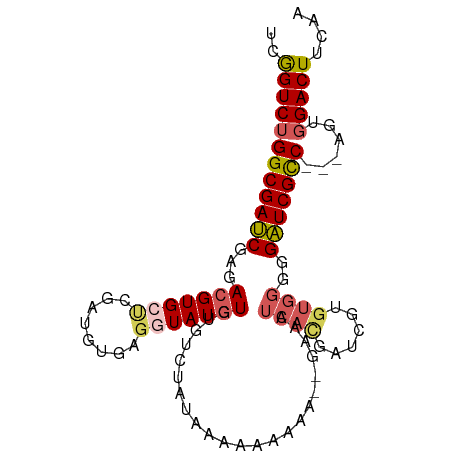
>dm3.chr3R 23119083 94 + 27905053 UCGGUCUGGCGAUCGAGACGUGCUCGAUGUGAGGUAUGUGUGUCUAUAAAAAAAAA------UCAUGAUCGUGUGGGGGAUCGCCGCCAGUGGACUUCAA ..(((..(((((((.(((((..(..............)..)))))...........------((((......))))..))))))))))............ ( -24.84, z-score = 0.14, R) >droAna3.scaffold_13340 4125516 93 - 23697760 CCAGUCUGGCGAGCGAGACGUGCCCGCAUGGAGGUAUUUCUAUAAAAGUGCCAUGAUCGA----CCAGUUUUCUGAUAGUUCGUC---ACUUGACUCAUG ..((((((((((((((..((((....))))..(((((((......)))))))....))..----.(((....)))...)))))))---)...)))).... ( -26.00, z-score = -0.95, R) >droEre2.scaffold_4820 5555409 95 - 10470090 CCGGUCUGGCGAUCGAGACGUGCUCGAUGUGAGGUAUGUUCGAGUUUGUCUAAUAA--AAAAUCACGAUCGAGUGGGGGAUCGUC---AGUGGACUUGGG ((((((((((((((.(((((.((((((............)))))).))))).....--....((((......))))..)))))))---...)))).))). ( -33.10, z-score = -2.24, R) >droSec1.super_13 1873113 95 + 2104621 UCGGUCUGGCGAUCGAGACGUGCUCGAUGUGAGUUAUGUGUGUCUAUAAAAAAAAA--GAAAUCACGAUCGUGUGGGGGAUCGCC---AGUGGACUUCAA ((.(.(((((((((.(((((..(..(((....)))..)..)))))...........--....(((((....)))))..)))))))---))).))...... ( -29.80, z-score = -2.05, R) >droSim1.chr3R 22889802 97 + 27517382 UCGGUCUGGCGAUCGAGACGUGCUCGAUGUGACUUAUGUGUGUCUAUAAAAAAAAAAAGAAAUCACGAUCGUGUGGGGGAUCGCC---CGUGGACUUCAA ..((((((((((((.(((((..(..............)..))))).................(((((....)))))..)))))))---...))))).... ( -26.94, z-score = -0.96, R) >consensus UCGGUCUGGCGAUCGAGACGUGCUCGAUGUGAGGUAUGUGUGUCUAUAAAAAAAAA__GAAAUCACGAUCGUGUGGGGGAUCGCC___AGUGGACUUCAA ..((((((((((((...(((((((........))))))).......................((((......))))..)))))))......))))).... (-17.30 = -18.14 + 0.84)
| Location | 23,119,083 – 23,119,177 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 71.97 |
| Shannon entropy | 0.49232 |
| G+C content | 0.49166 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -10.18 |
| Energy contribution | -10.42 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23119083 94 - 27905053 UUGAAGUCCACUGGCGGCGAUCCCCCACACGAUCAUGA------UUUUUUUUUAUAGACACACAUACCUCACAUCGAGCACGUCUCGAUCGCCAGACCGA ..........((((((..((((........))))....------............................((((((.....))))))))))))..... ( -20.10, z-score = -1.56, R) >droAna3.scaffold_13340 4125516 93 + 23697760 CAUGAGUCAAGU---GACGAACUAUCAGAAAACUGG----UCGAUCAUGGCACUUUUAUAGAAAUACCUCCAUGCGGGCACGUCUCGCUCGCCAGACUGG ....((((((((---(..((.(.(((((....))))----).).))....)))))..................((((((.......))))))..)))).. ( -21.90, z-score = -0.20, R) >droEre2.scaffold_4820 5555409 95 + 10470090 CCCAAGUCCACU---GACGAUCCCCCACUCGAUCGUGAUUUU--UUAUUAGACAAACUCGAACAUACCUCACAUCGAGCACGUCUCGAUCGCCAGACCGG ..........((---(.(((((...(((......))).....--.....((((...(((((............)))))...)))).))))).)))..... ( -19.30, z-score = -1.66, R) >droSec1.super_13 1873113 95 - 2104621 UUGAAGUCCACU---GGCGAUCCCCCACACGAUCGUGAUUUC--UUUUUUUUUAUAGACACACAUAACUCACAUCGAGCACGUCUCGAUCGCCAGACCGA ..........((---(((((((........(((.((((....--........................)))))))(((.....))))))))))))..... ( -21.89, z-score = -2.52, R) >droSim1.chr3R 22889802 97 - 27517382 UUGAAGUCCACG---GGCGAUCCCCCACACGAUCGUGAUUUCUUUUUUUUUUUAUAGACACACAUAAGUCACAUCGAGCACGUCUCGAUCGCCAGACCGA .....(((....---(((((((........(((.(((((((.......(((....))).......))))))))))(((.....)))))))))).)))... ( -22.54, z-score = -1.91, R) >consensus UUGAAGUCCACU___GGCGAUCCCCCACACGAUCGUGAUUUC__UUUUUUUUUAUAGACACACAUACCUCACAUCGAGCACGUCUCGAUCGCCAGACCGA .....(((........((((((........)))))).....................................(((((.....)))))......)))... (-10.18 = -10.42 + 0.24)

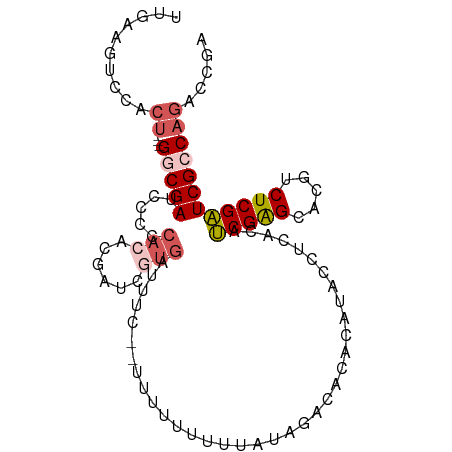
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:42 2011