| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,794,640 – 8,794,741 |
| Length | 101 |
| Max. P | 0.650914 |

| Location | 8,794,640 – 8,794,741 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.88 |
| Shannon entropy | 0.37760 |
| G+C content | 0.44111 |
| Mean single sequence MFE | -17.51 |
| Consensus MFE | -13.00 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8794640 101 - 23011544 AAAUCCGAGUUCCAAUUCCAAUU----------UCCCCCGAAAUGAGGCCC-AAAAUACCACUUUAUAUAAUACUGUUGACAUAUGUUAAUCAGGCGUACGAACACGCGCCG ......((((....))))..(((----------((....)))))..(((..-.....................((((((((....))))).)))((((......))))))). ( -17.00, z-score = -1.21, R) >droSim1.chr2L 8572781 100 - 22036055 AAAUCCGAGUUCCAAUUCCAA-U----------UCCCCCGAAAUGAGGCCC-AAAAUACCACUUUAUAUAAUACUGUUGACAUAUGUUAAUCAGGCGUACGAACACGCGCCG ......(((((........))-)----------))...........(((..-.....................((((((((....))))).)))((((......))))))). ( -16.60, z-score = -1.14, R) >droSec1.super_3 4266475 100 - 7220098 AAAUCCGAGUUCCAAUUCCAA-U----------UCCCCCGAAAUGAGGCCC-AAAAUACCACUUUAUAUAAUACUGUUGACAUAUGUUAAUCAGGCGUACGAACACGCGCCG ......(((((........))-)----------))...........(((..-.....................((((((((....))))).)))((((......))))))). ( -16.60, z-score = -1.14, R) >droYak2.chr2L 11438487 100 - 22324452 AAAUCCGAGUUCCAAUUCCAA-U----------UCCCCCGAAAUGAGGCCC-AAAAUACCACUUUAUAUAAUACUGUUGACAUAUGUUAAUCAGGCGUACGAACACGCGCCG ......(((((........))-)----------))...........(((..-.....................((((((((....))))).)))((((......))))))). ( -16.60, z-score = -1.14, R) >droEre2.scaffold_4929 9382757 100 - 26641161 AAAUCCGAGUUCCAAUUCCAA-U----------UCCCCCGAAAUGAGGCCC-AAAAUACCACUUUAUAUAAUACUGUUGACAUAUGUUAAUCAGGCGUACGAACACGCGCCG ......(((((........))-)----------))...........(((..-.....................((((((((....))))).)))((((......))))))). ( -16.60, z-score = -1.14, R) >droAna3.scaffold_12916 10455633 111 - 16180835 AAAUCCGUAUUCCGAUUCCAAUUUCGAUUCCGAUCCCCCGAAAUGAGGCCC-AAAAUACCACUUUAUAUAAUACUGUUGACAUAUGUUAAUCAGGCGUACGAACACGCGCCG ....................((((((............))))))..(((..-.....................((((((((....))))).)))((((......))))))). ( -18.70, z-score = -0.60, R) >dp4.chr4_group5 1583383 95 + 2436548 ------GGGACCGAAUUCCAAU-----------UCCCCAGAAAUGAGGCCCAAAAAUACCACUUUAUAUAAUACUGUUGACAUAUGUUAAUCAGGCGUACGAACACGCGCCG ------(((.((((((....))-----------))..((....)).)))))........................((((((....))))))..(((((........))))). ( -20.70, z-score = -1.73, R) >droPer1.super_5 1553259 81 + 6813705 --------------------AU-----------UCCCCAGAAAUGAGGCCCAAAAAUACCACUUUAUAUAAUACUGUUGACAUAUGUUAAUCAGGCGUACGAACACGCGCCG --------------------..-----------....((....)).(((........................((((((((....))))).)))((((......))))))). ( -16.70, z-score = -2.24, R) >droVir3.scaffold_12723 4782064 91 - 5802038 --------AACGCAAUUCCAA-------------GCCCAAAUGCCCAGCACCAAAAUACCACUUUAUAUAAUACUGUUGACAUAUGUUAAUCAGGCGCACGCACACGCGCCG --------...((........-------------)).((.(((..(((((.....(((........))).....))))).))).)).......(((((........))))). ( -13.80, z-score = -0.20, R) >droMoj3.scaffold_6500 10752518 92 - 32352404 ------AAGGCCUUGGCACAG--------------UCCAAAUGCCCAGGACCGAAAUACCACUUUAUAUAAUACUGUUGACAUAUGUUAAUCAGGCGCACGCACACGCGCCG ------..(((((.((((...--------------......)))).))).)).......................((((((....))))))..(((((........))))). ( -24.70, z-score = -1.92, R) >droGri2.scaffold_15126 7784313 91 - 8399593 --------AACGCAAUUCCAA------------UCCCAAAUGCCCAGGCCC-AAAAUACCACUUUAUAUAAUACUGUUGACAUAUGUUAAUCAGGCGCACGCACACGCGCCG --------.............------------........((....))..-.......................((((((....))))))..(((((........))))). ( -14.60, z-score = -0.40, R) >consensus AAAUCCGAGUUCCAAUUCCAA_U__________UCCCCCGAAAUGAGGCCC_AAAAUACCACUUUAUAUAAUACUGUUGACAUAUGUUAAUCAGGCGUACGAACACGCGCCG ..............................................(((........................((((((((....))))).)))(((........)))))). (-13.00 = -13.27 + 0.27)

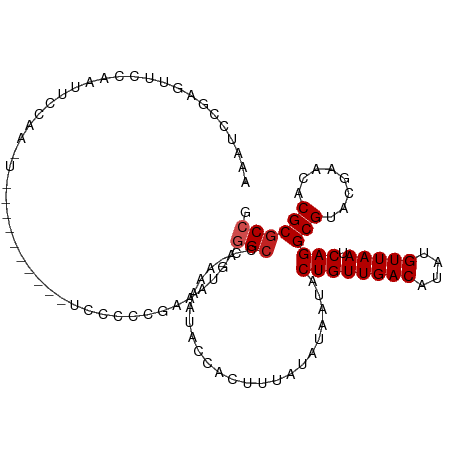

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:56 2011