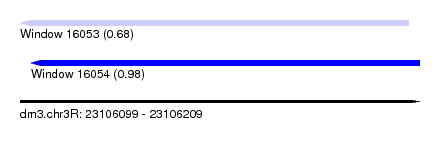
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,106,099 – 23,106,209 |
| Length | 110 |
| Max. P | 0.983777 |

| Location | 23,106,099 – 23,106,206 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.88 |
| Shannon entropy | 0.50722 |
| G+C content | 0.39963 |
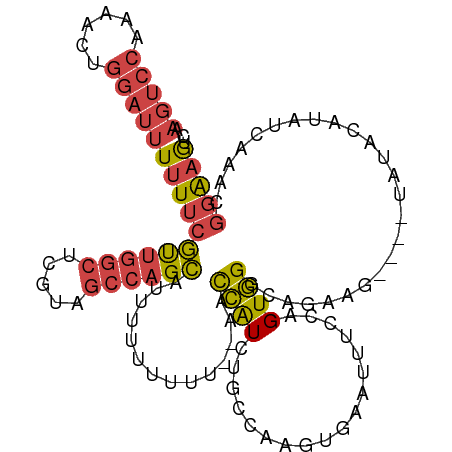
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -11.02 |
| Energy contribution | -11.77 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
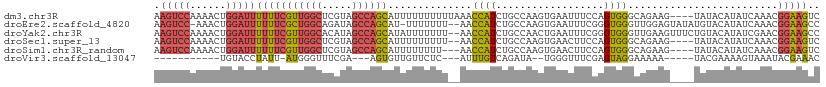
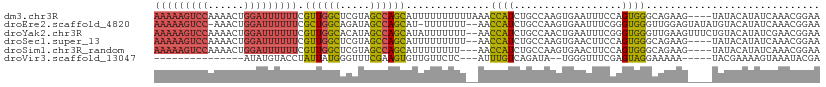
>dm3.chr3R 23106099 107 - 27905053 AAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAUUUUUUUUUUAAACCAUCUGCCAAGUGAAUUUCCAGUGGGCAGAAG----UAUACAUAUCAAACGGAAGUC (((((((....)))))))....((((((.....)))))).................((((((.(.((......)).).))))))..----............((....)). ( -28.30, z-score = -2.67, R) >droEre2.scaffold_4820 5542151 107 + 10470090 AAGUCC-AAACUGGAUUUUUUCGCUGGCAGAUAGCCAGCAU-UUUUUUU--AACCAUCUGCCAAGUGAAUUUCGGGUGGGUUGGAGUAUAUGUACAUAUCAAACGGAAGCC ((((((-.....))))))((((((((((.....))))))..-.......--..(((((..((..(......)..))..)).)))....................))))... ( -27.10, z-score = -0.84, R) >droYak2.chr3R 5444590 109 + 28832112 AAGUCCAAAACUGGAUUUUUUCGUUGGCACAUAGCCAGCAUAUUUUUUU--AACCAUCUGCCAACUGAAUUUCGGGUGGGUUGAAGUUUCUGUACAUAUCGAACGGAAGCC (((((((....)))))))....((((((.....)))))).......(((--(((((((((............))))).)))))))((((((((.(.....).)))))))). ( -31.50, z-score = -2.47, R) >droSec1.super_13 1860206 105 - 2104621 AAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAUUUUUUUUU--AACCAUCUGCCAAGUGAACUUCCAGUGGGCAGAAG----UAUACAUAUCAAACGGAAGUC (((((((....)))))))....((((((.....))))))..........--.....((((((.(.((......)).).))))))..----............((....)). ( -28.30, z-score = -2.53, R) >droSim1.chr3R_random 34468 104 - 1307089 AAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAUUUUUUUU---AACCAUCUGCCAAGUGAACUUCCAGUGGGCAGAAG----UAUACAUAUCAAACGGAAGUC (((((((....)))))))....((((((.....)))))).........---.....((((((.(.((......)).).))))))..----............((....)). ( -28.30, z-score = -2.49, R) >droVir3.scaffold_13047 7945277 86 - 19223366 -----------UGUACCUAUU-AUGGGUUUCGA---AGUGUUGUUCUC---AUUUGUCAGAUA--UGGGUUUCGAGUAGGAAAAA-----UACGAAAAGUAAAUACGAAAC -----------....((....-..))(((((..---.(((((.(((..---((((((((....--)))....)))))..))).))-----))).....((....))))))) ( -13.90, z-score = 0.22, R) >consensus AAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAUUUUUUUUU__AACCAUCUGCCAAGUGAAUUUCCAGUGGGCAGAAG____UAUACAUAUCAAACGGAAGUC .(((((......)))))(((((((((((.....))))))..............((((..................)))).........................))))).. (-11.02 = -11.77 + 0.75)
| Location | 23,106,102 – 23,106,209 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.59 |
| Shannon entropy | 0.53108 |
| G+C content | 0.37893 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.57 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
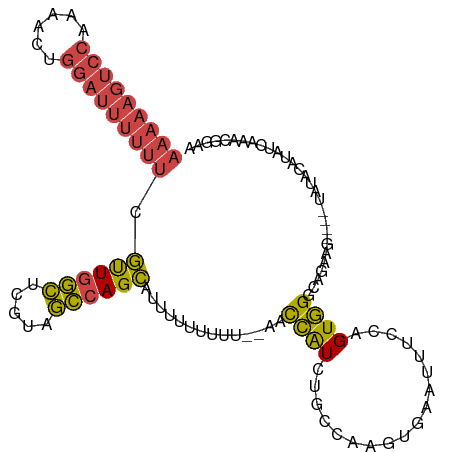
>dm3.chr3R 23106102 107 - 27905053 AAAAAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAUUUUUUUUUUAAACCAUCUGCCAAGUGAAUUUCCAGUGGGCAGAAG----UAUACAUAUCAAACGGAA ((((((((((....)))))))))).((((((.....))))))..............((.((((((.(.((......)).).)))))).(----....).........)).. ( -29.80, z-score = -3.20, R) >droEre2.scaffold_4820 5542154 107 + 10470090 AAAAAGUCC-AAACUGGAUUUUUUCGCUGGCAGAUAGCCAGCAU-UUUUUUU--AACCAUCUGCCAAGUGAAUUUCGGGUGGGUUGGAGUAUAUGUACAUAUCAAACGGAA (((((((((-.....))))))))).((((((.....))))))..-.......--..(((((..((..(......)..))..)).))).(((....)))............. ( -28.60, z-score = -1.69, R) >droYak2.chr3R 5444593 109 + 28832112 AAAAAGUCCAAAACUGGAUUUUUUCGUUGGCACAUAGCCAGCAUAUUUUUUU--AACCAUCUGCCAACUGAAUUUCGGGUGGGUUGAAGUUUCUGUACAUAUCGAACGGAA ((((((((((....)))))))))).((((((.....))))))......((((--(((((((((............))))).)))))))).((((((.(.....).)))))) ( -30.20, z-score = -2.41, R) >droSec1.super_13 1860209 105 - 2104621 AAAAAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAUUUUUUUUU--AACCAUCUGCCAAGUGAACUUCCAGUGGGCAGAAG----UAUACAUAUCAAACGGAA ((((((((((....)))))))))).((((((.....))))))..........--..((.((((((.(.((......)).).)))))).(----....).........)).. ( -29.80, z-score = -3.10, R) >droSim1.chr3R_random 34471 104 - 1307089 AAAAAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAUUUUUUUU---AACCAUCUGCCAAGUGAACUUCCAGUGGGCAGAAG----UAUACAUAUCAAACGGAA ((((((((((....)))))))))).((((((.....)))))).........---..((.((((((.(.((......)).).)))))).(----....).........)).. ( -29.80, z-score = -3.06, R) >droVir3.scaffold_13047 7945280 86 - 19223366 ---------------AUAUGUACCUAUUAUGGGUUUCGAAGUGUUGUUCUC---AUUUGUCAGAUA--UGGGUUUCGAGUAGGAAAAA-----UACGAAAAGUAAAUACGA ---------------...((((((......))........(((((.(((..---((((((((....--)))....)))))..))).))-----)))..........)))). ( -12.30, z-score = 0.96, R) >consensus AAAAAGUCCAAAACUGGAUUUUUUCGUUGGCUCGUAGCCAGCAUUUUUUUUU__AACCAUCUGCCAAGUGAAUUUCCAGUGGGCAGAAG____UAUACAUAUCAAACGGAA ((((((((((....)))))))))).((((((.....))))))..............((((..................))))............................. (-14.22 = -14.57 + 0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:36 2011