| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,080,785 – 23,080,880 |
| Length | 95 |
| Max. P | 0.971732 |

| Location | 23,080,785 – 23,080,880 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 67.25 |
| Shannon entropy | 0.68346 |
| G+C content | 0.55065 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -14.76 |
| Energy contribution | -14.73 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
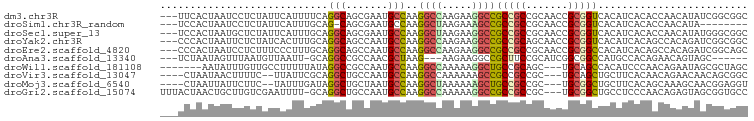
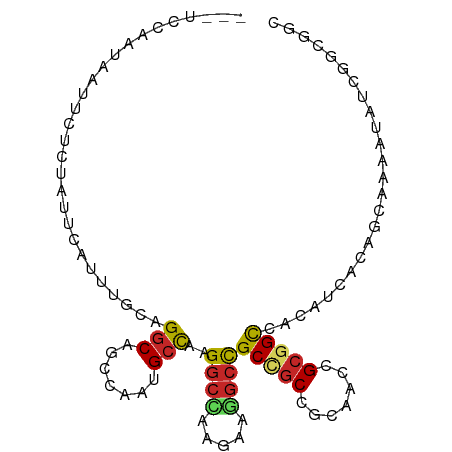
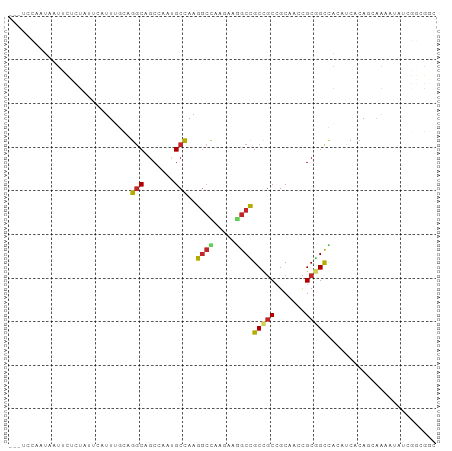
>dm3.chr3R 23080785 95 + 27905053 ---UUCACUAAUCCUCUAUUCAUUUUCAGGCAGCGAAUGCCAAGGCCAAGAAGGCCGCCGCCGCAACCGCGGUCACAUCACACCAACAUAUCGGCGGC ---.........................((((.....))))..((((.....))))((((((((....))(((........)))........)))))) ( -30.80, z-score = -3.18, R) >droSim1.chr3R_random 1080527 86 + 1307089 ---UCCACUAAUCCUCUAUUCAUUUGCAG-CAGCGAAUGCCAAGGCUAAGAAAGCCGCCGCCGCAACCGCGGUCACAUCACACCAACAUA-------- ---.................(((((((..-..)))))))....((((.....))))(..(((((....)))))..)..............-------- ( -19.80, z-score = -1.81, R) >droSec1.super_13 1835023 95 + 2104621 ---UCCACUAAUGCUCUAUUCAUUUGCAGGCAGCGAAUGCCAAGGCUAAGAAGGCCGCCGCCGCAACCGCGGUCACAUCACACCAACAUAUGGGCGGC ---........(((...........)))((((.....))))..((((.....))))((((((((....))(((........)))........)))))) ( -29.80, z-score = -0.91, R) >droYak2.chr3R 5419071 95 - 28832112 ---CCCACUAAUUCUCUAUCACUUUGCAGGCAGCCAAUGCCAAGGCCAAGAAGGCCGCCGCAGCAACCGCGGUCACAUCACAGCCACAGAUCGGCGGC ---.........................((((.....))))..((((.....))))(((((.((....))((((..............)))).))))) ( -27.04, z-score = -0.70, R) >droEre2.scaffold_4820 5516562 95 - 10470090 ---CCCACUAAUCCUCUUUCCCUUUGCAGGCAGCCAAUGCCAAGGCCAAGAAGGCCGCCGCCGCAACCGCGGCCACAUCACAGCCACAGAUCGGCAGC ---......................((.((((.....))))..((((.....)))))).(((((....))))).........(((.......)))... ( -29.30, z-score = -1.48, R) >droAna3.scaffold_13340 4086351 85 - 23697760 ---UCUAAUAGUUUAAUGUUAAUU-GCAGGCCGCCAACGCUAAG---AAGAAGGCCGCUUCCGCAUCGGCGGCCAUGCCACAGAACAGUAGC------ ---.......(((...((((....-((((((((((...(((...---.....))).((....))...))))))).))).....))))..)))------ ( -27.10, z-score = -1.82, R) >droWil1.scaffold_181108 4350117 88 - 4707319 -------AAUAUUUGUUGCCUUUUUAUAGGCCGCCAAUGCCAAGGCCAAAAAGGCUGCCGCAGC---UGCAGCCACAUCCCAACAGAAUAGCGCUAGC -------....(((((((..........((((...........)))).....((((((......---.))))))......)))))))...((....)) ( -24.70, z-score = -0.36, R) >droVir3.scaffold_13047 7917696 89 + 19223366 ----CUAAUAACUUUUC--UUAUUCGCAGGCUGCCAAUGCCAAGGCCAAAAAAGCCGCCGCCGC---UGCAGCUGCUUCACAACAGAACAACAGCGGC ----.............--......((.((((......((....))......)))))).(((((---((...(((........))).....))))))) ( -24.80, z-score = -0.75, R) >droMoj3.scaffold_6540 5875617 89 - 34148556 ----CUAAUUAUUCUUC--UAUUUGAUAGGCUGCUAAUGCCAAGGCUAAAAAAGCUGCCGCCGC---UGCGGCUGCUUCACAGCAAAGCAACGGAGGU ----.........((((--(..(((....((((.....((....)).....((((.(((((...---.))))).))))..))))....))).))))). ( -26.50, z-score = -0.27, R) >droGri2.scaffold_15074 6039827 94 - 7742996 UUUACUAACUGCUUGUCGAAUUUU-GCAGGCUGCCAAUGCCAAGGCCAAAAAGGCCGCCGCCGC---UGCGGCUGCCUCCCAACAGAGUAGCGGUGCC ..........((((((........-))))))............((((.....))))..((((((---(((..(((........))).))))))))).. ( -32.70, z-score = -0.35, R) >consensus ___UCCAAUAAUUCUCUAUUCAUUUGCAGGCAGCCAAUGCCAAGGCCAAGAAGGCCGCCGCCGCAACCGCGGCCACAUCACAGCAAAAUAUCGGCGGC ............................(((.......)))..((((.....))))(((((.......)))))......................... (-14.76 = -14.73 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:34 2011