
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,793,472 – 8,793,581 |
| Length | 109 |
| Max. P | 0.781420 |

| Location | 8,793,472 – 8,793,581 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.57 |
| Shannon entropy | 0.31716 |
| G+C content | 0.40542 |
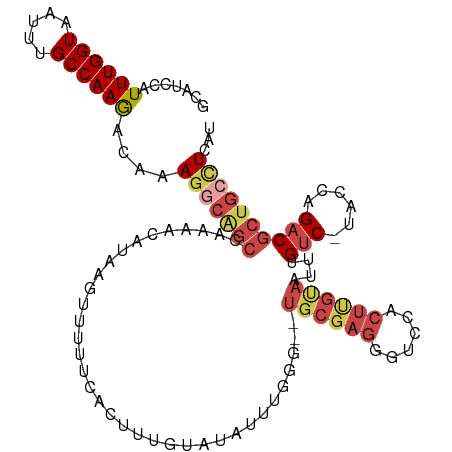
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -17.27 |
| Energy contribution | -18.88 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
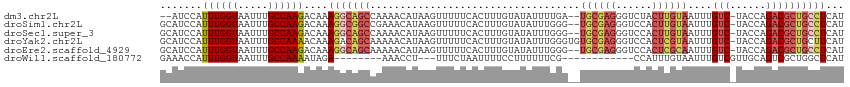
>dm3.chr2L 8793472 109 - 23011544 --AUCCAUUUGGUAAUUUGCCAAGACAAAGGCAGCCAAAACAUAAGUUUUUCACUUUGUAUAUUUUGA--UGCGAGGGUCUACUUGUAAUUUGUC-UACCAGACGCUGCCUCAU --.....((((((.....))))))....(((((((......((((((......((((((((......)--)))))))....)))))).....(((-.....))))))))))... ( -31.00, z-score = -3.08, R) >droSim1.chr2L 8571621 111 - 22036055 GCAUCCAUUUGGUAAUUUGCCAAGACAAAGGCGGCCGAAACAUAAGUUUUUCACUUUGUAUAUUUGGG--UGCGAGGGUCCACUUGUAAUUUGUC-UACCAGACGCUGCCUCAU .......((((((.....))))))....(((((((......((((((......((((((((......)--)))))))....)))))).....(((-.....))))))))))... ( -29.90, z-score = -1.01, R) >droSec1.super_3 4265308 111 - 7220098 GCAUCCAUUUGGUAAUUUGCCAAGACAAAGGCAGCCAAAACAUAAGUUUUUCACUUUGUAUAUUUGGG--UGCGAGGGUCCACUUGUAAUUUGUC-UACCAGACGCUGCCUCAU .......((((((.....))))))....(((((((......((((((......((((((((......)--)))))))....)))))).....(((-.....))))))))))... ( -30.60, z-score = -1.66, R) >droYak2.chr2L 11437291 113 - 22324452 GCAUCCAUUUGGUAAUUUGCCAAAACAAAGACAGCAAAAACAUAAGUUUUUCACUUUGUAUAUUUGGGUGUGCGAGGGUCCACUCGUAAUUUGUC-UACCAGACGCUGCUUCAU (((((((((((((.....))))))((((((...(.((((((....))))))).)))))).....)))))))(((((......))))).....(((-.....))).......... ( -29.50, z-score = -2.00, R) >droEre2.scaffold_4929 9381606 111 - 26641161 GCAUCCAUUUGGUAAUUUGCCAAGACAAAGGCAGCAAAAACAUAAGUUUUUCACUUUGUAUAUUUGGG--UGCGAGGGUCCACUCGCAAUUUGUC-UACCAGACGCUGCCUCAU .......((((((.....))))))....(((((((((((((....))))))...........(((((.--((((((......)))))).......-..))))).)))))))... ( -33.20, z-score = -2.62, R) >droWil1.scaffold_180772 77690 91 - 8906247 GAAACCAUUUGGUAAUUUGCCAAAAUAGA--------AAACCU---UUUCUAAUUUUCCUUUUUUCG------------CCAUUUGUAAUUUGUCGUUGCAGUCGCUGGCUCAU ((((...((((((.....)))))).((((--------((....---))))))..........))))(------------(((.(((((((.....)))))))....)))).... ( -18.90, z-score = -2.33, R) >consensus GCAUCCAUUUGGUAAUUUGCCAAGACAAAGGCAGCCAAAACAUAAGUUUUUCACUUUGUAUAUUUGGG__UGCGAGGGUCCACUUGUAAUUUGUC_UACCAGACGCUGCCUCAU .......((((((.....))))))....(((((((...................................((((((......))))))....(((......))))))))))... (-17.27 = -18.88 + 1.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:56 2011