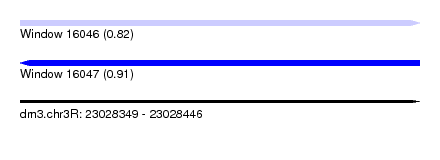
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,028,349 – 23,028,446 |
| Length | 97 |
| Max. P | 0.914228 |

| Location | 23,028,349 – 23,028,446 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 66.90 |
| Shannon entropy | 0.66518 |
| G+C content | 0.37872 |
| Mean single sequence MFE | -19.14 |
| Consensus MFE | -8.53 |
| Energy contribution | -8.62 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.822541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
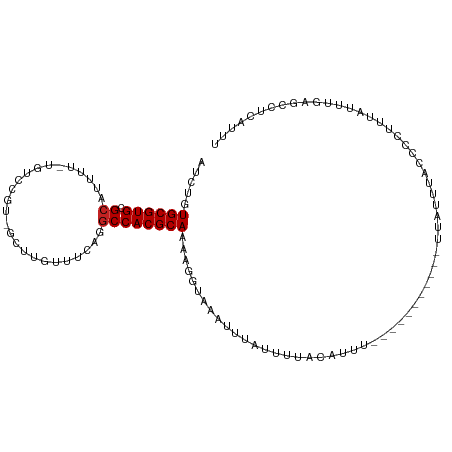
>dm3.chr3R 23028349 97 + 27905053 AAAUGAGGCUCAAAUGAAGGGGUAAAUAA------------AAUGUAAAAUAAAUUUACCUUUUGCGUGGCCUGAAACAAGCGACGGACA-AAAAAGCGCACGCACAGAU ...(.(((((...((((((((((((((..------------............))))))))))).)))))))).).....(((.(.....-.....)))).......... ( -19.54, z-score = -0.99, R) >droSim1.chr3R 22801774 97 + 27517382 AAAUGAGGCUCAAAUGAAGGGGUAAAUAA------------AAUGUAAAAUAAAUUUACCUUUUGCGUGGCCUGAAACAAGCGACGGACA-AAAAAGCGCACGCACAGAU ...(.(((((...((((((((((((((..------------............))))))))))).)))))))).).....(((.(.....-.....)))).......... ( -19.54, z-score = -0.99, R) >droSec1.super_13 1783124 97 + 2104621 AAAUGAGGCUCAAAUGAAGGGGUAAAUAA------------AAUGUAAAAUAAAUUUACCUUUUGCGUGGCCUGAAACAAGCGACGGACA-AAAAAGCGCACGCACAGAU ...(.(((((...((((((((((((((..------------............))))))))))).)))))))).).....(((.(.....-.....)))).......... ( -19.54, z-score = -0.99, R) >droYak2.chr3R 5367301 97 - 28832112 AAAUGAGUCUCAAAUGAAGGGGUAAAUAA------------AAUGUAAAAUGAAUUUACCUUUUGCGUGGCCUGAAACAAGCUACGGACA-AAAAAGCGCACGCACAGAU ......(((.......(((((((((((..------------.((.....))..))))))))))).((((((.((...)).))))))))).-.....((....))...... ( -21.40, z-score = -1.65, R) >droEre2.scaffold_4820 5461443 98 - 10470090 AAAUGAGGCUCAAAUGAAGGGUUAAAUAA------------AAUGUAAAAUAAAUUUACCUUUUGCGUGGCCUGAAACAAGCAACGGACACAAAAAGCGCACGCACAGAU ...(.((((.((..((((((((.((((..------------............))))))))))))..)))))).).....................((....))...... ( -15.94, z-score = 0.25, R) >droAna3.scaffold_12911 3965871 109 + 5364042 AAAUGAGCCUCAAAUAAGGUGGUAAAUAAGGCCUCAAAUAAAAUGUAAAAUGAAUUUACCUUUUGCGUGGCCAGAAACAAGCCAGCUACA-AAAAGCCUCACGCACAGAU ...((((.((......((.((((......((((.(...(((((.(((((.....))))).))))).).))))........)))).))...-...)).))))......... ( -20.44, z-score = -0.31, R) >dp4.chr2 8651482 107 + 30794189 AAAUGAGCUUCGAAUAAACAACUGGUAAAA--UACGAGUUAAAUGUUAAAUUAACGUACACUUUGCGUGGCUCGAAACCAACCAAAGACU-GAAAUGCGCACGCACAGAU ...((((((.((.......((((.(((...--))).))))..((((((...))))))........)).))))))..............((-(...(((....)))))).. ( -19.50, z-score = -0.57, R) >droPer1.super_0 2464977 107 + 11822988 AAAUGAGCUUCGAAUAAACAACUGGUAAAA--UACGAGUUAAAUGUUAAAUUAACGUACACUUUGCGUGGCUCGAAACCAACCAAAGACU-GAAAUGCGCACGCACAGAU ...((((((.((.......((((.(((...--))).))))..((((((...))))))........)).))))))..............((-(...(((....)))))).. ( -19.50, z-score = -0.57, R) >droVir3.scaffold_13047 8170492 75 + 19223366 ---------------AAAUGAGCC-UCAAA--UAAAGGGUAAAUA--AAAUAAAUUUACGUUUUGCGUGGCGUGAAA---------------AAAUGCGCACGCACAGAU ---------------.........-.....--.((((.((((((.--......)))))).))))(((((((((....---------------..)))).)))))...... ( -18.30, z-score = -2.16, R) >droMoj3.scaffold_6540 6127221 75 - 34148556 ---------------AAAUGAGCC-UCAAA--UAAAGGGUAAAUA--AAAUAAAUUUACGUUUUGCGUGGCGUGAAA---------------AAAUGCGCACGCACAGAU ---------------.........-.....--.((((.((((((.--......)))))).))))(((((((((....---------------..)))).)))))...... ( -18.30, z-score = -2.16, R) >droGri2.scaffold_15074 6299475 75 - 7742996 ---------------AAAUGAGCC-UCAUA--UAAAGGGUAAAUA--AAAUAAAUUUACGUUUUGCGUGGCGUGAAA---------------AAAUGCGCACGCACAGAU ---------------..(((....-.))).--.((((.((((((.--......)))))).))))(((((((((....---------------..)))).)))))...... ( -18.50, z-score = -2.13, R) >consensus AAAUGAGGCUCAAAUAAAGGGGUAAAUAA___________AAAUGUAAAAUAAAUUUACCUUUUGCGUGGCCUGAAACAAGC_ACGGACA_AAAAAGCGCACGCACAGAU ...............................................................((((((((.........................)).))))))..... ( -8.53 = -8.62 + 0.09)

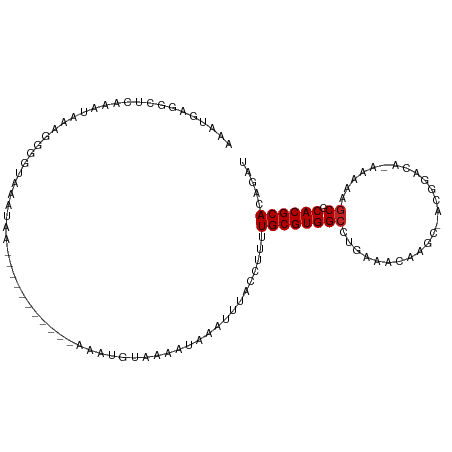
| Location | 23,028,349 – 23,028,446 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 66.90 |
| Shannon entropy | 0.66518 |
| G+C content | 0.37872 |
| Mean single sequence MFE | -17.99 |
| Consensus MFE | -9.21 |
| Energy contribution | -9.30 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23028349 97 - 27905053 AUCUGUGCGUGCGCUUUUU-UGUCCGUCGCUUGUUUCAGGCCACGCAAAAGGUAAAUUUAUUUUACAUU------------UUAUUUACCCCUUCAUUUGAGCCUCAUUU ........(.(.(((((((-((..(((.(((((...))))).))))))))(((((((..(........)------------..))))))).........))))).).... ( -17.40, z-score = -1.09, R) >droSim1.chr3R 22801774 97 - 27517382 AUCUGUGCGUGCGCUUUUU-UGUCCGUCGCUUGUUUCAGGCCACGCAAAAGGUAAAUUUAUUUUACAUU------------UUAUUUACCCCUUCAUUUGAGCCUCAUUU ........(.(.(((((((-((..(((.(((((...))))).))))))))(((((((..(........)------------..))))))).........))))).).... ( -17.40, z-score = -1.09, R) >droSec1.super_13 1783124 97 - 2104621 AUCUGUGCGUGCGCUUUUU-UGUCCGUCGCUUGUUUCAGGCCACGCAAAAGGUAAAUUUAUUUUACAUU------------UUAUUUACCCCUUCAUUUGAGCCUCAUUU ........(.(.(((((((-((..(((.(((((...))))).))))))))(((((((..(........)------------..))))))).........))))).).... ( -17.40, z-score = -1.09, R) >droYak2.chr3R 5367301 97 + 28832112 AUCUGUGCGUGCGCUUUUU-UGUCCGUAGCUUGUUUCAGGCCACGCAAAAGGUAAAUUCAUUUUACAUU------------UUAUUUACCCCUUCAUUUGAGACUCAUUU .(((.((((((.((((...-.((.....)).......))))))))))...(((((((............------------..)))))))..........)))....... ( -16.74, z-score = -0.73, R) >droEre2.scaffold_4820 5461443 98 + 10470090 AUCUGUGCGUGCGCUUUUUGUGUCCGUUGCUUGUUUCAGGCCACGCAAAAGGUAAAUUUAUUUUACAUU------------UUAUUUAACCCUUCAUUUGAGCCUCAUUU ....(((.(.(((((((((((((.....(((((...))))).)))))))))))................------------............((....))))).))).. ( -17.00, z-score = -0.37, R) >droAna3.scaffold_12911 3965871 109 - 5364042 AUCUGUGCGUGAGGCUUUU-UGUAGCUGGCUUGUUUCUGGCCACGCAAAAGGUAAAUUCAUUUUACAUUUUAUUUGAGGCCUUAUUUACCACCUUAUUUGAGGCUCAUUU ....(((.((((((((((.-.((((((((((.......))))).)).....(((((.....)))))....)))..)))))))))).)))..((((....))))....... ( -30.70, z-score = -2.44, R) >dp4.chr2 8651482 107 - 30794189 AUCUGUGCGUGCGCAUUUC-AGUCUUUGGUUGGUUUCGAGCCACGCAAAGUGUACGUUAAUUUAACAUUUAACUCGUA--UUUUACCAGUUGUUUAUUCGAAGCUCAUUU ....((((....))))...-.......(((.(((((((((.(((.....))).................(((((.(((--...))).)))))....))))))))).))). ( -17.20, z-score = 1.05, R) >droPer1.super_0 2464977 107 - 11822988 AUCUGUGCGUGCGCAUUUC-AGUCUUUGGUUGGUUUCGAGCCACGCAAAGUGUACGUUAAUUUAACAUUUAACUCGUA--UUUUACCAGUUGUUUAUUCGAAGCUCAUUU ....((((....))))...-.......(((.(((((((((.(((.....))).................(((((.(((--...))).)))))....))))))))).))). ( -17.20, z-score = 1.05, R) >droVir3.scaffold_13047 8170492 75 - 19223366 AUCUGUGCGUGCGCAUUU---------------UUUCACGCCACGCAAAACGUAAAUUUAUUU--UAUUUACCCUUUA--UUUGA-GGCUCAUUU--------------- .....((((((.((....---------------......))))))))....((((((......--.))))))((((..--...))-)).......--------------- ( -15.60, z-score = -2.31, R) >droMoj3.scaffold_6540 6127221 75 + 34148556 AUCUGUGCGUGCGCAUUU---------------UUUCACGCCACGCAAAACGUAAAUUUAUUU--UAUUUACCCUUUA--UUUGA-GGCUCAUUU--------------- .....((((((.((....---------------......))))))))....((((((......--.))))))((((..--...))-)).......--------------- ( -15.60, z-score = -2.31, R) >droGri2.scaffold_15074 6299475 75 + 7742996 AUCUGUGCGUGCGCAUUU---------------UUUCACGCCACGCAAAACGUAAAUUUAUUU--UAUUUACCCUUUA--UAUGA-GGCUCAUUU--------------- .....((((((.((....---------------......))))))))....((((((......--.))))))((((..--...))-)).......--------------- ( -15.60, z-score = -2.21, R) >consensus AUCUGUGCGUGCGCAUUUU_UGUCCGU_GCUUGUUUCAGGCCACGCAAAAGGUAAAUUUAUUUUACAUUU___________UUAUUUACCCCUUUAUUUGAGCCUCAUUU .....((((((.((.........................))))))))............................................................... ( -9.21 = -9.30 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:30 2011