| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,997,401 – 22,997,493 |
| Length | 92 |
| Max. P | 0.995643 |

| Location | 22,997,401 – 22,997,493 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 76.30 |
| Shannon entropy | 0.45614 |
| G+C content | 0.38386 |
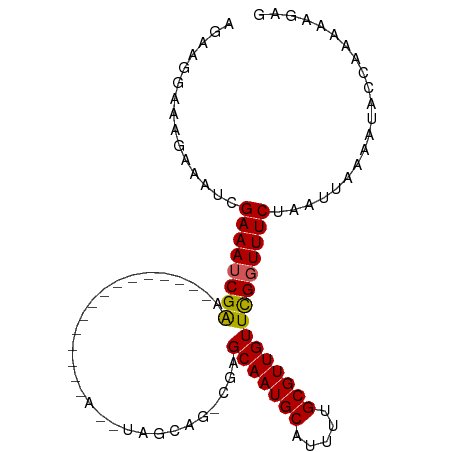
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -13.03 |
| Energy contribution | -12.86 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.995643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
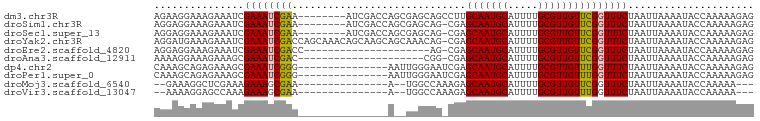
>dm3.chr3R 22997401 92 - 27905053 AGAAGGAAAGAAAUCGAAAUCGAA--------AUCGACCAGCGAGCAGCCUUGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACCAAAAAGAG ........((((((((((.(((..--------..)))...(((((....)))))(((((.....))))).)))))))))).................... ( -21.50, z-score = -1.64, R) >droSim1.chr3R 22793883 91 - 27517382 AGGAGGAAAGAAAUCGAAAUCGAA--------AUCGACCAGCGAGCAG-CGAGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACCAAAAAGAG ....((...((..(((....))).--------.))((((.((.....)-)(((((((((.....))))))))))))).............))........ ( -23.90, z-score = -2.34, R) >droSec1.super_13 1752506 91 - 2104621 AGGAGGAAAGAAAUCGAAAUCGAA--------AUCGACCAGCGAGCAG-CGAGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACCAAAAAGAG ....((...((..(((....))).--------.))((((.((.....)-)(((((((((.....))))))))))))).............))........ ( -23.90, z-score = -2.34, R) >droYak2.chr3R 5335922 99 + 28832112 AGGAUGAAAGAAAUCGAAAUCGACCAGCAAACAGCAAGCAGCAAACAG-CGAGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACCAAAAAGAG ........((((((((((..((((..(((((..(((.((.((.....)-)..))..)))..))))))))))))))))))).................... ( -23.20, z-score = -2.25, R) >droEre2.scaffold_4820 5430807 78 + 10470090 AGGAGGAAAGAAAUCGAAAUCGACC---------------------AG-CGAGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACCAAAAAGAG ........(((((((((..(((...---------------------..-)))(((((((.....)))))))))))))))).................... ( -19.30, z-score = -2.35, R) >droAna3.scaffold_12911 3935386 78 - 5364042 AAAAGGAAAGAAAGCGAAAUCGAC---------------------CGG-CGAGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACCAAAAAGAG ........(((((.((....))..---------------------...-((((((((((.....)))))))))).))))).................... ( -17.70, z-score = -1.53, R) >dp4.chr2 8605551 85 - 30794189 CAAAGCAGAGAAAGCGAAAUCGGG---------------AAUUGGGAAUCGAGCAAUGCAUUUUGCGUUGUUUGGUUUCUAAUUAAAAUACCAAAAAGAG ....((.......))......((.---------------((((((((((((((((((((.....))))))))))))))))))))......))........ ( -26.80, z-score = -4.58, R) >droPer1.super_0 2432980 85 - 11822988 CAAAGCAGAGAAAGCGAAAUCGGG---------------AAUUGGGAAUCGAGCAAUGCAUUUUGCGUUGUUUGGUUUCUAAUUAAAAUACCAAAAAGAG ....((.......))......((.---------------((((((((((((((((((((.....))))))))))))))))))))......))........ ( -26.80, z-score = -4.58, R) >droMoj3.scaffold_6540 6090950 78 + 34148556 --GAAAGGCUCGAAAGAAAGCGAA---------------A--UGGCCAAAGAGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACCAAAAA--- --....(((((....)).......---------------.--..)))...(((((((((.....)))))))))(((.............))).....--- ( -19.52, z-score = -2.08, R) >droVir3.scaffold_13047 8134047 78 - 19223366 --AAAAGGAGCCAAAGAAAGCGAA---------------A--UGGCCAAAGAGCAAUGCAUUUUGCGUUGUUUGGUUUCUAAUUAAAAUACCAAAAA--- --...((((((((((.((.(((((---------------(--((((......))....)))))))).)).)))))))))).................--- ( -22.40, z-score = -3.38, R) >consensus AGAAGGAAAGAAAUCGAAAUCGAA_______________A__UAGCAG_CGAGCAAUGCAUUUUGCGUUGUUCGGUUUCUAAUUAAAAUACCAAAAAGAG ...............((((((((.............................(((((((.....)))))))))))))))..................... (-13.03 = -12.86 + -0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:26 2011