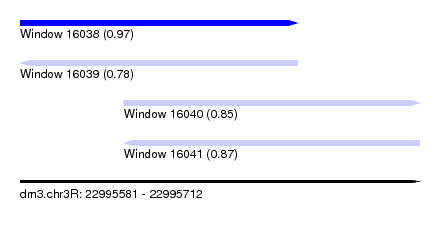
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,995,581 – 22,995,712 |
| Length | 131 |
| Max. P | 0.967471 |

| Location | 22,995,581 – 22,995,672 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 59.48 |
| Shannon entropy | 0.77157 |
| G+C content | 0.50695 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -14.45 |
| Energy contribution | -15.32 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
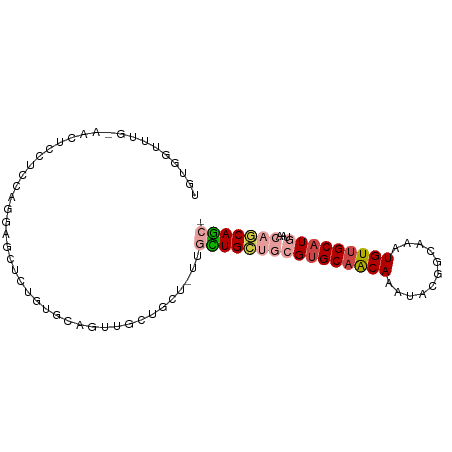
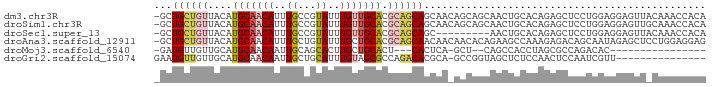
>dm3.chr3R 22995581 91 + 27905053 UGUGGUUUGUAACUCCUCCAGGAGCUCUGUGCAGUUGCUGCUGUUGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGC- .(..(((((((.((((....)))).....)))))..))..).(((((((.(((((((((((...........))))))))))).)))))))- ( -35.30, z-score = -1.75, R) >droSim1.chr3R 22792088 91 + 27517382 UGUGGUUUGCAACUCCUCCAGGAGCUCUGUGCAGUUGCUGCUGUUGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGC- .(..(((((((.((((....)))).....)))))..))..).(((((((.(((((((((((...........))))))))))).)))))))- ( -37.30, z-score = -1.92, R) >droSec1.super_13 1750721 82 + 2104621 UGUGGUUUGUAACUCCUCCAGGAGCUCUGUGCAGUU---------GCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGC- .......((((.((((....)))).....))))(((---------((((.(((((((((((...........))))))))))).)))))))- ( -31.10, z-score = -1.93, R) >droAna3.scaffold_12911 3933484 91 + 5364042 CUCCUCCAGGAGCUCUAUUGCUGUCUCUUUGGCUUCUGUGUUGUUGUUGCUGCGUGCAGCAAAUACAGCAAAUGUUGCAUGUAACAGCAGC- ((((....)))).....(((((((..(....((..(..((((((..((((((....))))))..))))))...)..))..)..))))))).- ( -31.20, z-score = -1.75, R) >droMoj3.scaffold_6540 6089170 69 - 34148556 ----------------GUGUCUGGCGCUAGGUGGCUG--AGC-UGAGUG---AGUGCAGCAAGUGCUGCAAUUGUUGCAUGCAACAACAUC- ----------------........((((.(((.....--.))-).))))---..((((((....)))))).((((((....))))))....- ( -20.70, z-score = 1.17, R) >droGri2.scaffold_15074 6260653 76 - 7742996 ---------------AACGAUUGGAGUUGGAGAGCUACCGGC-UGCGUGUCUGGCGCUACAAAUGCAGCAAUUGUUGCAUGCAACAACAUUC ---------------......(((((((....)))).)))((-(((((...((......)).)))))))..((((((....))))))..... ( -21.00, z-score = -0.00, R) >consensus UGUGGUUUG_AACUCCUCCAGGAGCUCUGUGCAGUUGCUGCU_UUGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAGC_ .............................................((((((((((((((((...........)))))))))...))))))). (-14.45 = -15.32 + 0.86)

| Location | 22,995,581 – 22,995,672 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 59.48 |
| Shannon entropy | 0.77157 |
| G+C content | 0.50695 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -9.62 |
| Energy contribution | -10.43 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22995581 91 - 27905053 -GCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAACAGCAGCAACUGCACAGAGCUCCUGGAGGAGUUACAAACCACA -((((((((....(((((((...........))))))).))))))))....((((...))))....((((((....)))))).......... ( -33.40, z-score = -2.37, R) >droSim1.chr3R 22792088 91 - 27517382 -GCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAACAGCAGCAACUGCACAGAGCUCCUGGAGGAGUUGCAAACCACA -((((((((....(((((((...........))))))).)))))))).......((((((.(.(((.....)))..).))))))........ ( -34.90, z-score = -2.08, R) >droSec1.super_13 1750721 82 - 2104621 -GCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGC---------AACUGCACAGAGCUCCUGGAGGAGUUACAAACCACA -((((((((....(((((((...........))))))).))))))))---------..........((((((....)))))).......... ( -29.60, z-score = -2.35, R) >droAna3.scaffold_12911 3933484 91 - 5364042 -GCUGCUGUUACAUGCAACAUUUGCUGUAUUUGCUGCACGCAGCAACAACAACACAGAAGCCAAAGAGACAGCAAUAGAGCUCCUGGAGGAG -..((((((((((.(((.....))))))..((((((....)))))).....................)))))))......((((....)))) ( -24.40, z-score = -0.36, R) >droMoj3.scaffold_6540 6089170 69 + 34148556 -GAUGUUGUUGCAUGCAACAAUUGCAGCACUUGCUGCACU---CACUCA-GCU--CAGCCACCUAGCGCCAGACAC---------------- -...(((((((....)))))))((((((....))))))..---......-(((--.((....))))).........---------------- ( -17.80, z-score = -0.01, R) >droGri2.scaffold_15074 6260653 76 + 7742996 GAAUGUUGUUGCAUGCAACAAUUGCUGCAUUUGUAGCGCCAGACACGCA-GCCGGUAGCUCUCCAACUCCAAUCGUU--------------- ((.((..((((...((.((....(((((.((((......))))...)))-))..)).))....))))..)).))...--------------- ( -17.40, z-score = 0.42, R) >consensus _GCUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAA_ACCAGCAACUGCACAGAGCCACUGGAGGAGUU_CAAACCACA ...((((((....(((((((..((...))..))))))).))))))............................................... ( -9.62 = -10.43 + 0.81)
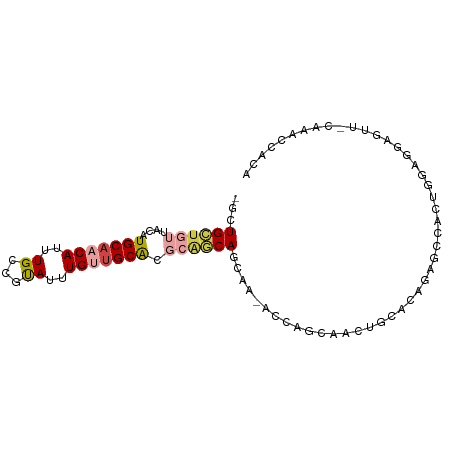
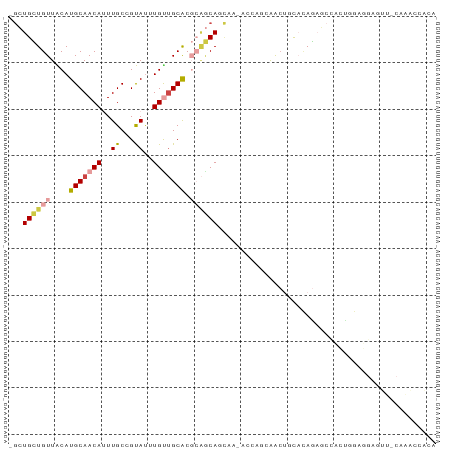
| Location | 22,995,615 – 22,995,712 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.77 |
| Shannon entropy | 0.56337 |
| G+C content | 0.42890 |
| Mean single sequence MFE | -31.03 |
| Consensus MFE | -13.19 |
| Energy contribution | -12.43 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
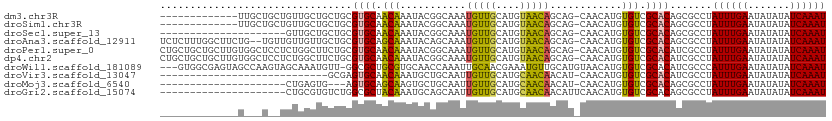
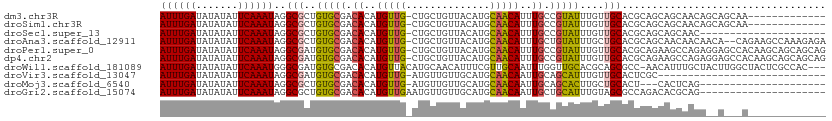
>dm3.chr3R 22995615 97 + 27905053 -------------UUGCUGCUGUUGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAG-CAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAU -------------..((((.((((((((((((((((((((...........)))))))))...))))))-)))))((.....))))))....((((((.......)))))) ( -34.90, z-score = -2.16, R) >droSim1.chr3R 22792122 97 + 27517382 -------------UUGCUGCUGUUGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAG-CAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAU -------------..((((.((((((((((((((((((((...........)))))))))...))))))-)))))((.....))))))....((((((.......)))))) ( -34.90, z-score = -2.16, R) >droSec1.super_13 1750754 89 + 2104621 ---------------------GUUGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAG-CAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAU ---------------------(((((((((((((((((((...........)))))))))...))))))-))))..((((......))))..((((((.......)))))) ( -31.70, z-score = -2.57, R) >droAna3.scaffold_12911 3933507 108 + 5364042 UCUCUUUGGCUUCUG--UGUUGUUGUUGCUGCGUGCAGCAAAUACAGCAAAUGUUGCAUGUAACAGCAG-CAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAU .......(((..(((--(((((((((((((((((((((((...........)))))))))...))))))-)))))......)))))).))).((((((.......)))))) ( -39.00, z-score = -3.06, R) >droPer1.super_0 2431239 110 + 11822988 CUGCUGCUGCUUGUGGCUCCUCUGGCUUCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAG-CAACAUGUGUCGCACAUCGCCUAUUUGAAUAUAUAUCAAAU .((((((((...(.((((.....)))).)(((((((((((...........))))))))))).))))))-))..(((((...))))).....((((((.......)))))) ( -32.50, z-score = -1.25, R) >dp4.chr2 8603856 110 + 30794189 CUGCUGCUGCUUGUGGCUCCUCUGGCUUCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAG-CAACAUGUGUCGCACAUCGCCUAUUUGAAUAUAUAUCAAAU .((((((((...(.((((.....)))).)(((((((((((...........))))))))))).))))))-))..(((((...))))).....((((((.......)))))) ( -32.50, z-score = -1.25, R) >droWil1.scaffold_181089 5394450 107 + 12369635 ---GUGGCGAGUAGCCAAGUAGCAAAUGUU-GGCGCUGCGUGCAACCAAAUUGCAACGAAAUGUUGCAUGUAACAUGUGUCGCACAUCGCCCAUUUGAAUAUAUAUCAAAU ---(.((((((((((....((((....)))-)..)))))((((.((((...((((((.....)))))).......)).)).)))).))))))((((((.......)))))) ( -31.20, z-score = -0.83, R) >droVir3.scaffold_13047 8132445 82 + 19223366 ----------------------------GCGAGUGCAACAAAUGCUGCAAUUGUUGCAUGCAACAACAU-CAACAUGUGUCGCACAUCGCCUAUUUGAAUAUAUAUCAAAU ----------------------------((((((((.(((.(((.((...((((((....))))))...-)).))).))).)))).))))..((((((.......)))))) ( -24.50, z-score = -2.45, R) >droMoj3.scaffold_6540 6089193 86 - 34148556 ---------------------CUGAGUG---AGUGCAGCAAGUGCUGCAAUUGUUGCAUGCAACAACAU-CAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAU ---------------------(((.(((---(.((((((....)))))).((((((....))))))...-.........)))).))).....((((((.......)))))) ( -23.00, z-score = -0.14, R) >droGri2.scaffold_15074 6260679 90 - 7742996 ---------------------CUGCGUGUCUGGCGCUACAAAUGCAGCAAUUGUUGCAUGCAACAACAUUCAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAU ---------------------..........((((((....(((((((....)))))))((.(((.(((.....)))))).))..)))))).((((((.......)))))) ( -26.10, z-score = -1.40, R) >consensus _____________U___U__UGUGGCUGCUGCGUGCAACAAAUACGGCAAAUGUUGCAUGUAACAGCAG_CAACAUGUGUCGCACAGCGCCUAUUUGAAUAUAUAUCAAAU ................................((((.(((...........(((((.......))))).........))).)))).......((((((.......)))))) (-13.19 = -12.43 + -0.76)
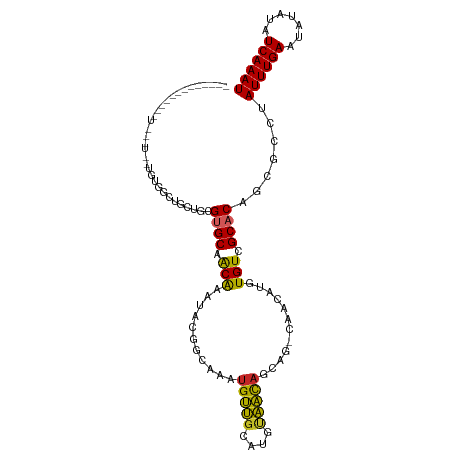
| Location | 22,995,615 – 22,995,712 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.77 |
| Shannon entropy | 0.56337 |
| G+C content | 0.42890 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -12.40 |
| Energy contribution | -12.38 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
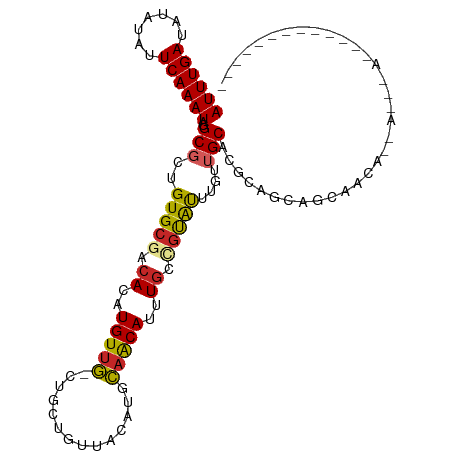
>dm3.chr3R 22995615 97 - 27905053 AUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUG-CUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAACAGCAGCAA------------- ((((((.......))))))....(((((........(((((-(((((((....(((((((...........))))))).)))))))))))))))))..------------- ( -36.80, z-score = -2.77, R) >droSim1.chr3R 22792122 97 - 27517382 AUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUG-CUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAACAGCAGCAA------------- ((((((.......))))))....(((((........(((((-(((((((....(((((((...........))))))).)))))))))))))))))..------------- ( -36.80, z-score = -2.77, R) >droSec1.super_13 1750754 89 - 2104621 AUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUG-CUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAAC--------------------- ((((((.......)))))).(.(((...))).)....((((-(((((((....(((((((...........))))))).)))))))))))--------------------- ( -29.90, z-score = -1.84, R) >droAna3.scaffold_12911 3933507 108 - 5364042 AUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUG-CUGCUGUUACAUGCAACAUUUGCUGUAUUUGCUGCACGCAGCAACAACAACA--CAGAAGCCAAAGAGA ((((((.......)))))).(((.(((((.......(((((-(((((((((((.(((.....)))))))......))).)))))))))....))--)))..)))....... ( -33.30, z-score = -1.81, R) >droPer1.super_0 2431239 110 - 11822988 AUUUGAUAUAUAUUCAAAUAGGCGAUGUGCGACACAUGUUG-CUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGAAGCCAGAGGAGCCACAAGCAGCAGCAG ((((((.......)))))).(((.(((((...))))))))(-((((((((...(((((((...........))))))).((....)).............))))))))).. ( -30.30, z-score = -0.27, R) >dp4.chr2 8603856 110 - 30794189 AUUUGAUAUAUAUUCAAAUAGGCGAUGUGCGACACAUGUUG-CUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGAAGCCAGAGGAGCCACAAGCAGCAGCAG ((((((.......)))))).(((.(((((...))))))))(-((((((((...(((((((...........))))))).((....)).............))))))))).. ( -30.30, z-score = -0.27, R) >droWil1.scaffold_181089 5394450 107 - 12369635 AUUUGAUAUAUAUUCAAAUGGGCGAUGUGCGACACAUGUUACAUGCAACAUUUCGUUGCAAUUUGGUUGCACGCAGCGCC-AACAUUUGCUACUUGGCUACUCGCCAC--- ((((((.......)))))).(((((.(((((((.((.((....((((((.....)))))))).))))))))).....(((-((..........)))))...)))))..--- ( -34.10, z-score = -2.61, R) >droVir3.scaffold_13047 8132445 82 - 19223366 AUUUGAUAUAUAUUCAAAUAGGCGAUGUGCGACACAUGUUG-AUGUUGUUGCAUGCAACAAUUGCAGCAUUUGUUGCACUCGC---------------------------- ((((((.......))))))..((((.((((((((.((((((-..(((((((....)))))))..)))))).))))))))))))---------------------------- ( -35.40, z-score = -4.91, R) >droMoj3.scaffold_6540 6089193 86 + 34148556 AUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUG-AUGUUGUUGCAUGCAACAAUUGCAGCACUUGCUGCACU---CACUCAG--------------------- ((((((.......))))))....((.(((((((((((....-))).))))))))))......((((((....))))))..---.......--------------------- ( -25.90, z-score = -1.27, R) >droGri2.scaffold_15074 6260679 90 + 7742996 AUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUGAAUGUUGUUGCAUGCAACAAUUGCUGCAUUUGUAGCGCCAGACACGCAG--------------------- ((((((.......)))))).(((((.(((((((((((.....))).)))))))))).......(((((....))))))))..........--------------------- ( -30.00, z-score = -1.88, R) >consensus AUUUGAUAUAUAUUCAAAUAGGCGCUGUGCGACACAUGUUG_CUGCUGUUACAUGCAACAUUUGCCGUAUUUGUUGCACGCAGCAGCAACA__A___A_____________ ((((((.......))))))..(((..(((((.((..(((((..((......))..)))))..)).)))))....))).................................. (-12.40 = -12.38 + -0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:25 2011