
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,955,661 – 22,955,763 |
| Length | 102 |
| Max. P | 0.858801 |

| Location | 22,955,661 – 22,955,763 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 62.11 |
| Shannon entropy | 0.69653 |
| G+C content | 0.52350 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -14.34 |
| Energy contribution | -13.73 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.48 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.858801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
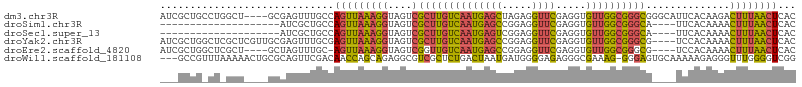
>dm3.chr3R 22955661 102 + 27905053 AUCGCUGCCUGGCU----GCGAGUUUGCCAGUUAAAGGUAGUCGCUUGUCAAUGAGCUAGAGGUUCGAGGUGUUGGCGGGCGGGCAUUCACAAGACUUUAACUCAC ...(((((((.(((----(((....)).))))...)))))))(((((((((((((((.....)))).....)))))))))))........................ ( -29.60, z-score = 0.62, R) >droSim1.chr3R 22775433 82 + 27517382 --------------------AUCGCUGCCAGUUAAAGGUAGUCGCUUGUCAAUGAGCCGGAGGUUCGAGGUGUUGGCGGGCA----UUCACAAAACUUUAACUCAC --------------------...((((((.......)))))).(((((((((((((((...))))).....)))))))))).----.................... ( -23.30, z-score = -0.90, R) >droSec1.super_13 1718643 82 + 2104621 --------------------AUCGCUGCCAGUUAAAGGUAGUCGCUUGUCAAUGAGUCGGAGGUUCGAGGUGUUGGCGGGCA----UUCACAAAACUUUAACUCAC --------------------...((((((.......)))))).(((((((((((..((((....))))..))))))))))).----.................... ( -24.60, z-score = -1.85, R) >droYak2.chr3R 5300249 102 - 28832112 AUCGCUGGCUCGCUCGUUGCGAGUUUGCGAGUUAAAGGUAGUCGCUUGUCAAUGAGCCGGAGGUUCGAGGUGUUGGCGGGCG----UCCACAAAACUUUAACUCAC ...((..((((((.....))))))..))(((((((((((.(.((((((((((((((((...))))).....)))))))))))----.).))....))))))))).. ( -44.20, z-score = -3.78, R) >droEre2.scaffold_4820 5395578 97 - 10470090 AUCGCUGGCUCGCU----GCUAGUUUGC-AGUUAAAGGUAGUCGGUUGUCAAUGAGCCGGAGGUUCGAGGUGUUGGCGGGCG----UCCACAAAACUUUAACUCAC ...((((((.....----)))))).((.-((((((((......((((.......))))(((.(((((.........))))).----)))......)))))))))). ( -29.60, z-score = -0.45, R) >droWil1.scaffold_181108 1462044 102 + 4707319 ---GCCGUUUAAAAACUGCGCAGUUCGACAACCAGCAGAGGCGUCGCUCUGACUAAUGAUGGGGAGAGGGCGAAAG-GGGAGUGCAAAAAGAGGGUUUGGGGUCGG ---.((.(((.....((((...(((....)))..))))...((((.((((..(((....)))..))))))))))).-))((.(.((((.......)))).).)).. ( -25.10, z-score = 1.12, R) >consensus ___GCUG_CU_GCU____GCAAGUUUGCCAGUUAAAGGUAGUCGCUUGUCAAUGAGCCGGAGGUUCGAGGUGUUGGCGGGCG____UUCACAAAACUUUAACUCAC .............................(((((((((....)((((((((((((((.....)))).....))))))))))..............))))))))... (-14.34 = -13.73 + -0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:19 2011