
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,953,455 – 22,953,558 |
| Length | 103 |
| Max. P | 0.627362 |

| Location | 22,953,455 – 22,953,558 |
|---|---|
| Length | 103 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 56.25 |
| Shannon entropy | 0.92931 |
| G+C content | 0.44855 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -7.74 |
| Energy contribution | -7.93 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
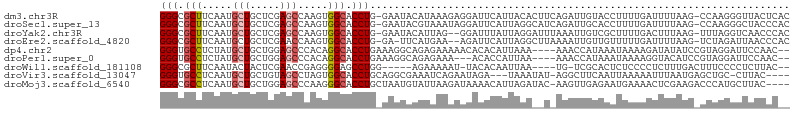
>dm3.chr3R 22953455 103 + 27905053 GGGCGCUUCAAUGCUGCUCGAGCCAAGUGGCACCUG-GAAUACAUAAAGAGGAUUCAUUACACUUCAGAUUGUACCUUUUGAUUUUAAG-CCAAGGGUUACUCAC ((((((......)).)))).((((...((((...((-((((.....((((((......((((........)))))))))).)))))).)-)))..))))...... ( -24.00, z-score = -0.31, R) >droSec1.super_13 1716438 103 + 2104621 GGGCGCUUCAAUGCUGCUCGAGCCAAGUGGCACCUG-GAAUACGUAAAUAGGAUUCAUUAGGCAUCAGAUUGCACCUUUUGAUUUUAAG-CCAAGGGCUACCCAC ((((((......)).)))).((((...((((.((((-...........))))..(((..(((((......)))..))..)))......)-)))..))))...... ( -28.80, z-score = -0.61, R) >droYak2.chr3R 5297966 101 - 28832112 GGGCGCUUCAAUGCUGCUCGAGCCAAGUGGCACCUG-GAAUACAUUAG--GGAUUUAUUAGGAUUUAAAUUGUCGCUUUUGACUUUAAG-UUUAGGUCAACCCAC ((((((......)).)))).....(((((((.((((-(......))))--)((((((........)))))))))))))(((((((....-...)))))))..... ( -25.30, z-score = -0.64, R) >droEre2.scaffold_4820 5393377 100 - 10470090 GGGCGCUUCAAUGCUGCUCGAACCAAGUGGCACCUG-GA-UUCAUGAA--AGAUUCAUUAGGCUUAAAAUUGUUGUUUUUGAUUUUAAG-UCUAGAUUAACCCAC (((....((..(((..((.......))..)))..((-((-((......--.)))))).((((((((((((((.......))))))))))-))))))....))).. ( -25.90, z-score = -1.63, R) >dp4.chr2 8561470 99 + 30794189 GGGUGCCUCUAUGCUGCUGGAGCCCACAGGCACCUGAAAGGCAGAGAAAAACACACAUUAAA----AAACCAUAAAUAAAAGAUAUAUCCGUAGGAUUCCAAC-- (((((..(((.((((.(.((.(((....))).)).)...)))).)))....)))........----....................((((...)))).))...-- ( -18.80, z-score = -0.21, R) >droPer1.super_0 2390369 96 + 11822988 GGGUGCCUCUAUGCUGCUGGAGCCCACAGGCACCUGAAAGGCAGAGAAA---ACACCAUUAA----AAACCAUAAAUAAAAGGUACAUCCGUAGGAUUCCAAC-- .((((..(((.((((.(.((.(((....))).)).)...)))).)))..---.)))).....----...............((...((((...)))).))...-- ( -23.70, z-score = -0.97, R) >droWil1.scaffold_181108 1459735 92 + 4707319 GGGCGCUUCAAUACUACUCGAACCGAGGGGAGCCUGG-----AGAAAAAU-UACACAAUUAA----UG-UCGCACUCUCCCCUCUUUGACUUUCCCCUCUUAC-- (((.(.(((..........))).)((((((((..((.-----.((...((-((......)))----).-)).))..))))))))..........)))......-- ( -21.80, z-score = -1.50, R) >droVir3.scaffold_13047 9833325 96 + 19223366 GGGUGCCUCAAUGCUGCUGUAGCCUAGUGGCACCUGCAGGCGAAAUCAGAAUAGA---UAAAUAU-AGGCUUCAAUUAAAAAUUUAAUGAGCUGC-CUUAC---- (((((((.(...((((...))))...).)))))))..((((...(((......))---)......-.(((((..(((((....))))))))))))-))...---- ( -25.60, z-score = -0.76, R) >droMoj3.scaffold_6540 24602556 100 - 34148556 GGGCGCCUCAAUGCUGCUGGAGCCCAAGGGCACCUGCUAAUGUAUUAAGAUAAAACAUUAGAUAC-AAGUUGAGAAUGAAAACUCGAAGACCCAUGCUUAC---- ((((.((.((....))..)).))))..((((.....(((((((.(((...))).)))))))....-...(((((........))))).).)))........---- ( -24.50, z-score = -1.33, R) >consensus GGGCGCUUCAAUGCUGCUCGAGCCAAGUGGCACCUG_GAAUAAAUAAAAAAGAUACAUUAAA__U_AAAUUGUAAAUUAAAAUUUUAAG_CCAAGGUUUACAC__ (((.(((.....(((.....))).....))).)))...................................................................... ( -7.74 = -7.93 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:18 2011