| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,909,417 – 22,909,511 |
| Length | 94 |
| Max. P | 0.972843 |

| Location | 22,909,417 – 22,909,511 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 57.78 |
| Shannon entropy | 0.76260 |
| G+C content | 0.65655 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -18.50 |
| Energy contribution | -19.65 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.972843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22909417 94 + 27905053 GAGUCUUGGCGGUGCCGGACCCACCGAUGGUUGGUGGUGCUGUUGUGGCAGAUGU--UGUGGCUGCGGCAGUGGUUGAGGCUGCGGCUGAGGAGCU-------------- (..(((..((.(((((.((((((((((...))))))..(((((((..((......--....))..)))))))))))..))).)).))..)))..).-------------- ( -41.70, z-score = -2.10, R) >droSim1.chr3R_random 1041599 94 + 1307089 AAGUCUUGGCGGUGCCGGACCCACCGAUGGUUGGUGGUGCUGUUGUGGCAGAUGC--UGUGGCUGCGGCAGUGGUUGGGGCUGCGGCUGAGGAGCU-------------- .(((((..((.(((((....(((((((...))))))).((.((..((((....))--))..)).)))))).).))..)))))..((((....))))-------------- ( -40.80, z-score = -1.13, R) >droYak2.chr3R 5251719 94 - 28832112 GAGUCUUGGCGGUGCCGGACCCACCGAAGGCUGGUGGUGCUGCUGUGGCAAAUGU--UGUGGUUGCGGCAGCGGUUGAGACUGUGGCUGUGGAGCU-------------- .(((((..((.(((((....((((((.....)))))).((.((((..((....))--..)))).)))))).).))..)))))..((((....))))-------------- ( -40.80, z-score = -1.84, R) >droEre2.scaffold_4820 5347782 100 - 10470090 GAGUCUUGGCGGUGCCGGACCCACCGACGGCUGGUGGUGCUGUUGUGGCAGAUGU--UGUGGUUGCGGCAGCGGUUGAGGCUGUGGCUGUGGUUGAGGAGCG-------- .(((((..((.((((((.(.(((((((((((.......))))))).(((....))--))))).).))))).).))..)))))...(((.(......).))).-------- ( -40.30, z-score = -0.83, R) >apiMel3.Group4 4547981 84 + 10796202 --------ACGGC-UCGUAGAGAGCCGUGGACGGCGUGACCGGGACGUAGGCCGG----UUGCUGGGCCGUU--CUGUAACCGCUGCCUCUCAAUUGGG----------- --------(((((-((.....)))))))((((((((..(((((........))))----)..)...))))))--)...........((........)).----------- ( -36.80, z-score = -1.05, R) >triCas2.ChLG2 6340341 107 - 12900155 GGGCGGGUCUAGCGUAGGACACUGGGGCGGGUCUUGGGGCUGGUCUGGGGGCUGGGGCGUAGCUGCG--AGCCGCUGGGGC-GUAGCUUUUAGGCGCUGGGCGAGGCGAG .(.(..(((((((((..((..((.((((.((((....)))).)))).))(((((.(.(.((((.(..--..).)))).).)-.))))).))..)))))))))..).)... ( -46.80, z-score = -0.81, R) >consensus GAGUCUUGGCGGUGCCGGACCCACCGAUGGCUGGUGGUGCUGUUGUGGCAGAUGU__UGUGGCUGCGGCAGCGGUUGAGGCUGCGGCUGUGGAGCU______________ .((((...(((((.......((((((.....))))))((((((((((((............))))))))))))......)))))))))...................... (-18.50 = -19.65 + 1.15)

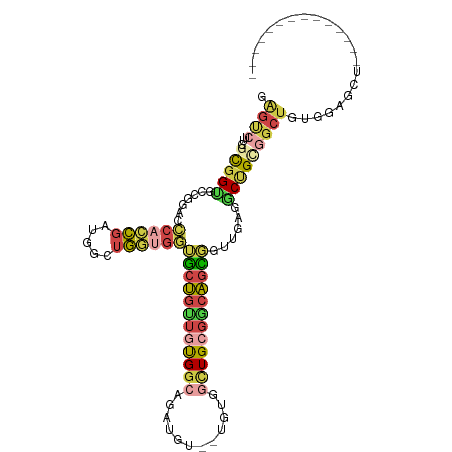
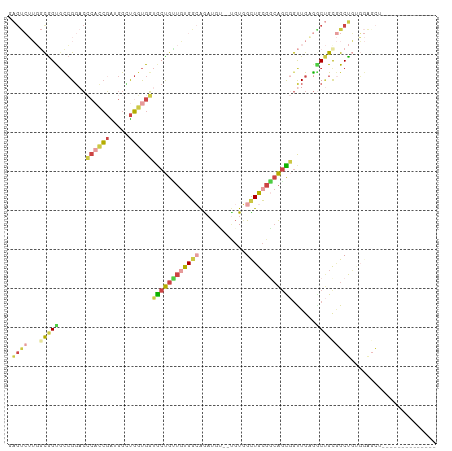
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:17 2011