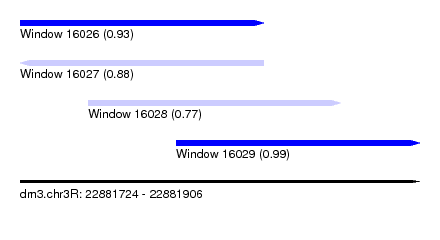
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,881,724 – 22,881,906 |
| Length | 182 |
| Max. P | 0.994458 |

| Location | 22,881,724 – 22,881,835 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.37 |
| Shannon entropy | 0.43607 |
| G+C content | 0.40419 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -18.22 |
| Energy contribution | -19.60 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
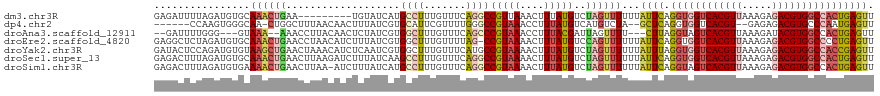
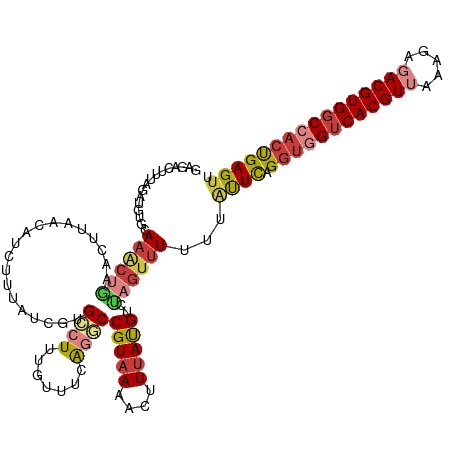
>dm3.chr3R 22881724 111 + 27905053 GAGAUUUUAGAUGUGCAAACUGAA---------UGUAUCAUGCCUUUGUUUCAGGCCGUUAAACUUUAUGUCUAGUUUUUUAUUCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUU ((((..(((((((((.....(((.---------....))).((((.......))))..........)))))))))..))))((((((.((((((((((.....)))))))))))))))). ( -32.80, z-score = -2.87, R) >dp4.chr2 8488264 109 + 30794189 ------CCAAGUGGGCAA-CUGGCUUUAACAACUUUAUCGUGCAUUCGUUUUGGGCCGUAAACCUUUAUGUCAUGUCUA--GCUCAGGUGGUCACGU--GAGAGACGUGCCCAAUGAGUU ------.....((((((.-.((((..(((....(((((.((.((.......)).)).)))))...))).))))))))))--(((((..(((.(((((--.....))))).))).))))). ( -28.40, z-score = 0.17, R) >droAna3.scaffold_12911 3800384 110 + 5364042 --GAUUUUGGG---GUAAA--AAACCUUACAACUCUAUCGUGGCUUUGUUUCAGCCCGUAAACCUUUACGAUUAGUUUU---CUUAGGUAGUCACGUUAAAGAUACGUGGCCACUGAGUU --.....((((---((...--..)))))).(((((....((((((.(((.((.....((((....))))(((((.((..---...)).)))))........)).))).)))))).))))) ( -27.20, z-score = -1.16, R) >droEre2.scaffold_4820 5319588 119 - 10470090 GAGGCUCUAGAUGUGCAAACUGAACCUAACAUCUUUAUCGUGGCUUUGUUUUAG-CCGUAAAACUUUAUGUCCAGUUUUUUAUUCAGGUGGUCACGUUAAAGAGACGUGGCCCCUGAGUU ...((.(.....).))((((((.((.(((....(((((...((((.......))-)))))))...))).)).))))))...(((((((.(((((((((.....)))))))))))))))). ( -36.10, z-score = -2.79, R) >droYak2.chr3R 5223598 120 - 28832112 GAUACUCCAGAUGUGUAAGCUGAACUAAACAUCUCAAUCGUGGCUUUGUUUCAUGCCGUAAAACUUUAUGUCUAGUUUUUUAUUUAGGUGGUCACGUUAAAGAGACGUGGCCACCGAGUU ...((((....((.(((((..((((((.((((.....(..((((..........))))..)......)))).))))))))))).))((((((((((((.....)))))))))))))))). ( -35.90, z-score = -3.15, R) >droSec1.super_13 1644241 120 + 2104621 GAGACUUUAGAUGUGCAAACUGAACUUAAGAUCUUUAUCAAGCCUUUGUUUCAGGCCGUAAAACUUUAUGUCUAGUUUUUUAUUCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUU .((((...((...(((...(((((...(((..(((....))).)))...)))))...)))...))....))))........((((((.((((((((((.....)))))))))))))))). ( -32.80, z-score = -2.02, R) >droSim1.chr3R 22707354 119 + 27517382 GAGACUUUAGAUGUGAAAACUGAACUUAA-AUCUUUAUCAUGCCUUUGUUUCAGGCCGUAAAACUUUAUGUCUAGUUUUUUAUUCAGGUAGUCACGUUAAAGAGACGUGGCCACUGAGUU ..(((((..((...((((((((.((.(((-(..(((((...((((.......)))).)))))..)))).)).))))))))...)).(((.((((((((.....)))))))).)))))))) ( -30.60, z-score = -2.11, R) >consensus GAGACUUUAGAUGUGCAAACUGAACUUAACAUCUUUAUCGUGCCUUUGUUUCAGGCCGUAAAACUUUAUGUCUAGUUUUUUAUUCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUU ................((((((...................((((.......))))(((((....)))))..))))))...((((.((((((((((((.....)))))))))))))))). (-18.22 = -19.60 + 1.38)
| Location | 22,881,724 – 22,881,835 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.37 |
| Shannon entropy | 0.43607 |
| G+C content | 0.40419 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -13.96 |
| Energy contribution | -15.36 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22881724 111 - 27905053 AACUCAGUGGCCACGUCUCUUUAACGUGACCACCUGAAUAAAAAACUAGACAUAAAGUUUAACGGCCUGAAACAAAGGCAUGAUACA---------UUCAGUUUGCACAUCUAAAAUCUC ......((((.(((((.......))))).))))((((((.......(((((.....)))))...((((.......)))).......)---------)))))................... ( -24.50, z-score = -2.63, R) >dp4.chr2 8488264 109 - 30794189 AACUCAUUGGGCACGUCUCUC--ACGUGACCACCUGAGC--UAGACAUGACAUAAAGGUUUACGGCCCAAAACGAAUGCACGAUAAAGUUGUUAAAGCCAG-UUGCCCACUUGG------ ....((.((((((((((((((--(.((....)).)))).--.))))..........((((((((((......((......)).....)))))..))))).)-.))))))..)).------ ( -26.30, z-score = -0.53, R) >droAna3.scaffold_12911 3800384 110 - 5364042 AACUCAGUGGCCACGUAUCUUUAACGUGACUACCUAAG---AAAACUAAUCGUAAAGGUUUACGGGCUGAAACAAAGCCACGAUAGAGUUGUAAGGUUU--UUUAC---CCCAAAAUC-- (((((.((((.(((((.......))))).)))).....---.......(((((...(.....).((((.......))))))))).)))))....(((..--...))---)........-- ( -25.40, z-score = -1.25, R) >droEre2.scaffold_4820 5319588 119 + 10470090 AACUCAGGGGCCACGUCUCUUUAACGUGACCACCUGAAUAAAAAACUGGACAUAAAGUUUUACGG-CUAAAACAAAGCCACGAUAAAGAUGUUAGGUUCAGUUUGCACAUCUAGAGCCUC ...(((((((.(((((.......))))).)).))))).....(((((((((.(((..(((((.((-((.......))))....)))))...))).)))))))))................ ( -32.30, z-score = -2.63, R) >droYak2.chr3R 5223598 120 + 28832112 AACUCGGUGGCCACGUCUCUUUAACGUGACCACCUAAAUAAAAAACUAGACAUAAAGUUUUACGGCAUGAAACAAAGCCACGAUUGAGAUGUUUAGUUCAGCUUACACAUCUGGAGUAUC .(((((((((.(((((.......))))).))))).........((((((((((..((((....(((..........)))..))))...))))))))))...............))))... ( -33.00, z-score = -3.30, R) >droSec1.super_13 1644241 120 - 2104621 AACUCAGUGGCCACGUCUCUUUAACGUGACCACCUGAAUAAAAAACUAGACAUAAAGUUUUACGGCCUGAAACAAAGGCUUGAUAAAGAUCUUAAGUUCAGUUUGCACAUCUAAAGUCUC ...(((((((.(((((.......))))).))).)))).....(((((..((.(((..((((..(((((.......))))).....))))..))).))..)))))................ ( -27.40, z-score = -2.00, R) >droSim1.chr3R 22707354 119 - 27517382 AACUCAGUGGCCACGUCUCUUUAACGUGACUACCUGAAUAAAAAACUAGACAUAAAGUUUUACGGCCUGAAACAAAGGCAUGAUAAAGAU-UUAAGUUCAGUUUUCACAUCUAAAGUCUC ...(((((((.(((((.......))))).))).))))....((((((..((.((((.(((((((((((.......)))).)).))))).)-))).))..))))))............... ( -26.80, z-score = -2.30, R) >consensus AACUCAGUGGCCACGUCUCUUUAACGUGACCACCUGAAUAAAAAACUAGACAUAAAGUUUUACGGCCUGAAACAAAGGCACGAUAAAGAUGUUAAGUUCAGUUUGCACAUCUAAAGUCUC ......((((.(((((.......))))).))))(((((...((((((........)))))).((((((.......)))).))..............)))))................... (-13.96 = -15.36 + 1.39)
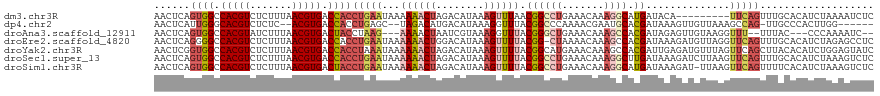
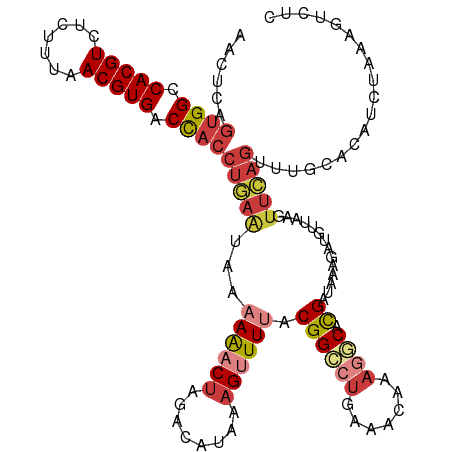
| Location | 22,881,755 – 22,881,870 |
|---|---|
| Length | 115 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.43 |
| Shannon entropy | 0.33494 |
| G+C content | 0.40206 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -17.44 |
| Energy contribution | -18.77 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22881755 115 + 27905053 UGCCUUUGUUUCAGGCCGUUAAACUUUAUGUCUAGUUUUUUAUUCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUUCAUCGUUAAAGUAAACUAAGCCACCAUAUUCGCAA----- (((...(((....(((.(((..((((((......(......((((((.((((((((((.....))))))))))))))))....)..)))))).)))...)))...)))...))).----- ( -28.90, z-score = -2.01, R) >dp4.chr2 8488297 116 + 30794189 UGCAUUCGUUUUGGGCCGUAAACCUUUAUGUCAUGUCUAGC--UCAGGUGGUCACGU--GAGAGACGUGCCCAAUGAGUUCAUCAUAAAACUAAAUUAUGCCGUCAUAUUCGAGAUUCUG ((.((((((..((((((((((....)))))..((((((..(--(((.((....)).)--)))))))))))))))))))).))........((....((((....))))....))...... ( -26.70, z-score = -0.35, R) >droAna3.scaffold_12911 3800417 117 + 5364042 UGGCUUUGUUUCAGCCCGUAAACCUUUACGAUUAGUUUUCU---UAGGUAGUCACGUUAAAGAUACGUGGCCACUGAGUUCAUCGUUAAAGUAAAUAAUGCCGUCAUUUUAGCCAUUCUG (((((..((..(.((........(((((((((.......((---(((...(((((((.......)))))))..)))))...)))).)))))........)).)..))...)))))..... ( -27.09, z-score = -1.96, R) >droEre2.scaffold_4820 5319628 118 - 10470090 UGGCUUUGUUUUA-GCCGUAAAACUUUAUGUCCAGUUUUUUAUUCAGGUGGUCACGUUAAAGAGACGUGGCCCCUGAGUUCAUUGUUACAGUAAACCACGCCACCAUAUUCGCAAUUCA- ((((...((((((-....))))))....(((.((((.....(((((((.(((((((((.....))))))))))))))))..))))..))).........))))................- ( -33.10, z-score = -3.16, R) >droYak2.chr3R 5223638 120 - 28832112 UGGCUUUGUUUCAUGCCGUAAAACUUUAUGUCUAGUUUUUUAUUUAGGUGGUCACGUUAAAGAGACGUGGCCACCGAGUUCAUUGUUAAAGUAAACUAAGCCACCAUAUUCUUAAAUCAG ((((((.(((((.......((((((........)))))).......((((((((((((.....))))))))))))...............).)))).))))))................. ( -34.50, z-score = -4.02, R) >droSec1.super_13 1644281 120 + 2104621 AGCCUUUGUUUCAGGCCGUAAAACUUUAUGUCUAGUUUUUUAUUCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUUCAUCGUUAAAGUAAACUAAGCCACCAUAUUCGCAAUUCGG .((...(((....((((((((....)))))..((((((...((((((.((((((((((.....)))))))))))))))).............)))))).)))...)))...))....... ( -29.29, z-score = -1.87, R) >droSim1.chr3R 22707393 120 + 27517382 UGCCUUUGUUUCAGGCCGUAAAACUUUAUGUCUAGUUUUUUAUUCAGGUAGUCACGUUAAAGAGACGUGGCCACUGAGUUCAUCGUUAAAGUAAACUAAGCCACCAUAUUCGCAAUUCGG (((...(((....((((((((....)))))..((((((...((((((...((((((((.....))))))))..)))))).............)))))).)))...)))...)))...... ( -24.79, z-score = -0.75, R) >consensus UGCCUUUGUUUCAGGCCGUAAAACUUUAUGUCUAGUUUUUUAUUCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUUCAUCGUUAAAGUAAACUAAGCCACCAUAUUCGCAAUUCAG .............((((((((....)))))................((((((((((((.....))))))))))))........................))).................. (-17.44 = -18.77 + 1.33)
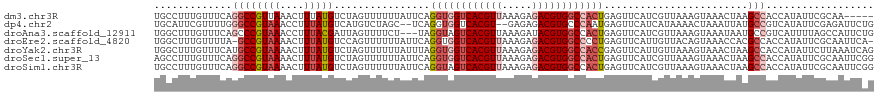
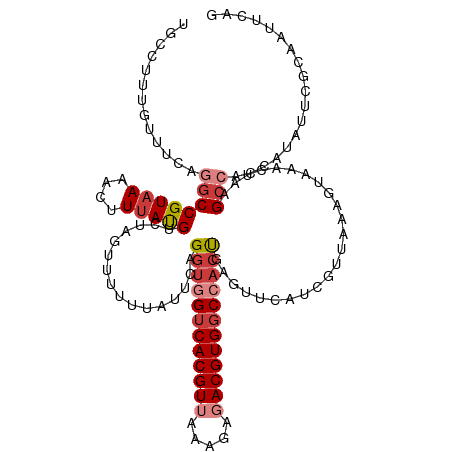
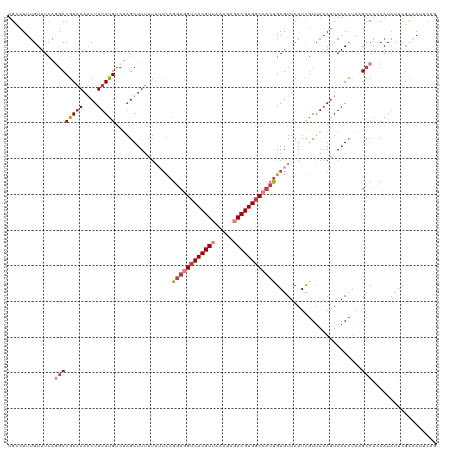
| Location | 22,881,795 – 22,881,906 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.40 |
| Shannon entropy | 0.49175 |
| G+C content | 0.41725 |
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.71 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.994458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
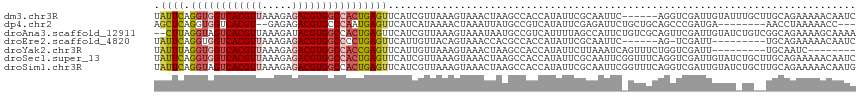
>dm3.chr3R 22881795 111 + 27905053 UAUUCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUUCAUCGUUAAAGUAAACUAAGCCACCAUAUUCGCAAUUC------AGGUCGAUUGUAUUUGCUUGCAGAAAAACAAUC ...((((.((((((((((.....))))))))))))))(((..((....(((((((.....(.(((.............------.))).)......)))))))...))..))).... ( -28.54, z-score = -2.36, R) >dp4.chr2 8488335 104 + 30794189 AGCUCAGGUGGUCACGU--GAGAGACGUGCCCAAUGAGUUCAUCAUAAAACUAAAUUAUGCCGUCAUAUUCGAGAUUCUGCUGCAGCCCGAUGA--------AACCUAAAAACC--- .(((((..(((.(((((--.....))))).))).)))))(((((......((....((((....))))....)).....((....))..)))))--------............--- ( -23.50, z-score = -1.13, R) >droAna3.scaffold_12911 3800456 115 + 5364042 --CUUAGGUAGUCACGUUAAAGAUACGUGGCCACUGAGUUCAUCGUUAAAGUAAAUAAUGCCGUCAUUUUAGCCAUUCUGUCGCAGUUCGAUUGUAUCUGUCGGCAGAAAAGCAAAA --(((((...(((((((.......)))))))..)))))......((((((((.............))))))))..(((((((((((...........))).))))))))........ ( -29.72, z-score = -2.45, R) >droEre2.scaffold_4820 5319667 101 - 10470090 UAUUCAGGUGGUCACGUUAAAGAGACGUGGCCCCUGAGUUCAUUGUUACAGUAAACCACGCCACCAUAUUCGCAAUUC------AG-UCGAUU---------UGCAGAAAAACAAUC .(((((((.(((((((((.....))))))))))))))))..((((((...((.....)).........(((((((.((------..-..)).)---------))).))).)))))). ( -31.40, z-score = -4.44, R) >droYak2.chr3R 5223678 100 - 28832112 UAUUUAGGUGGUCACGUUAAAGAGACGUGGCCACCGAGUUCAUUGUUAAAGUAAACUAAGCCACCAUAUUCUUAAAUCAGUUUCUGGUCGAUU---------UGCAAUC-------- ......((((((((((((.....))))))))))))...............(((((.....(.((((.(..((......))..).)))).).))---------)))....-------- ( -27.10, z-score = -3.13, R) >droSec1.super_13 1644321 117 + 2104621 UAUUCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUUCAUCGUUAAAGUAAACUAAGCCACCAUAUUCGCAAUUCGGUUUCAGGUCGAUUGUAUCUGCUUGCAGAAAAACAAUC .((((((.((((((((((.....))))))))))))))))................(((((((................))))).))...((((((.((((....))))...)))))) ( -32.79, z-score = -2.91, R) >droSim1.chr3R 22707433 117 + 27517382 UAUUCAGGUAGUCACGUUAAAGAGACGUGGCCACUGAGUUCAUCGUUAAAGUAAACUAAGCCACCAUAUUCGCAAUUCGGUUUCAGGUCGAUUGUAUCUGCUUGCAGAAAAACAAUG .((((((...((((((((.....))))))))..))))))................(((((((................))))).))....(((((.((((....))))...))))). ( -25.69, z-score = -0.90, R) >consensus UAUUCAGGUGGUCACGUUAAAGAGACGUGGCCACUGAGUUCAUCGUUAAAGUAAACUAAGCCACCAUAUUCGCAAUUC_GUUUCAGGUCGAUUGUAU_UG__UGCAGAAAAACAAUC .((((.((((((((((((.....)))))))))))))))).............................................................................. (-16.20 = -16.71 + 0.51)
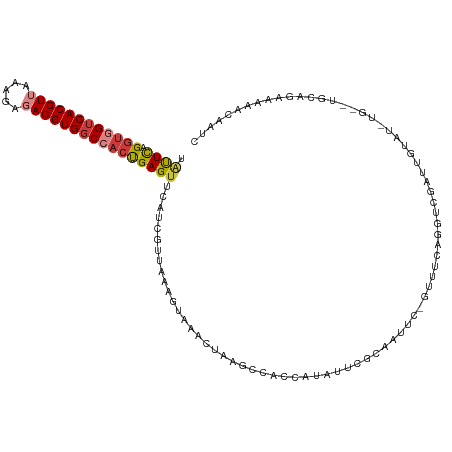
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:15 2011