
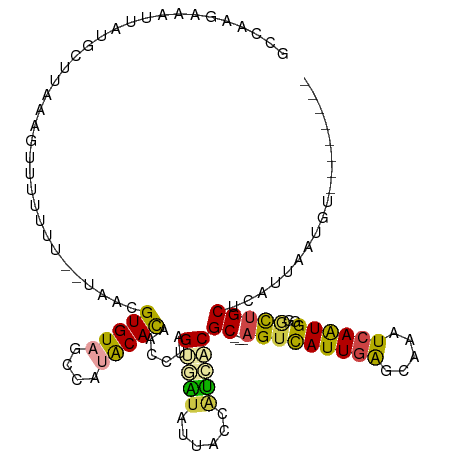
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,861,963 – 22,862,073 |
| Length | 110 |
| Max. P | 0.723529 |

| Location | 22,861,963 – 22,862,072 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.24 |
| Shannon entropy | 0.46040 |
| G+C content | 0.38458 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -14.37 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22861963 109 + 27905053 GCCAAAGUCUUACGCUUCAAGUUUUUUUUUUAACGUGUAGCCAUACACAACAUAGUGAUAUUACCAUCACGC---AGUCAUUGAGAAAAUCAAUGAGGCUGCUCAUUAAUGU-------- (((..........((...................(((((....)))))......(((((......)))))))---..(((((((.....)))))))))).............-------- ( -24.40, z-score = -2.22, R) >droAna3.scaffold_12911 3777340 115 + 5364042 ACCAAUAAACCAUUUAAAGAAUUUUCUC-----UGUGUAUCCACGCAUAACUGGGAGGUGUUCUCGCCUCGCCUGGGUUAUUCGGUUCAUCAAUGACUUGGCUCACUAAUGAGGUUUGCU .....((((((.................-----(((((....)))))....(((((((((....))))))(((.((((((((.((....)))))))))))))...)))....)))))).. ( -28.40, z-score = -0.91, R) >droEre2.scaffold_4820 5294956 106 - 10470090 GCCAAAGUAUUAUGUUUAGAGAUUUGUU--UAACGUGUAGCCAAGCACAACAUAGUAAUAUUACCUUCUCGC---AGUCAUUGAGCAAGUCAAUG-GACUGCUCAUUAAUGU-------- .......(((((((((((((......))--))..((((......))))))))))))).............((---(((((((((.....))))).-))))))..........-------- ( -22.20, z-score = -0.87, R) >droYak2.chr3R 5200431 107 - 28832112 ACCAAGGUAUUAUGCUUAAAGUUUUUUU--UAACGUGUAGCCAUGCACAACCUAGUGAUAUUACCAUCUCGC---AGUCAUUGAGCAAAUCAGUGGGGCUGCUCAUUAAUGU-------- .....((((.((((((....(((.....--.)))(((((....))))).....))).))).)))).....((---(((((((((.....))))))..)))))..........-------- ( -25.00, z-score = -0.75, R) >droSec1.super_13 1625669 107 + 2104621 GCCAAGAAAUUAUGGUAAAAGUUUUUUU--UAACGUGUAGCCAUACACAACCUAGUUGUAUUACCAUGACGC---AGUCAUUGAGCAAAUCAAUGGGGCUGCUCAUUAAUGU-------- .....((..(((((((((..(((.....--.)))(((((....)))))............))))))))).((---(((((((((.....))))))..)))))))........-------- ( -27.70, z-score = -1.99, R) >droSim1.chr3R 22687639 105 + 27517382 GCCAAGAAAUUAUGCUUAAUGUUUUUU----AACGUGUAGCCAUACACAACCUAGUUAUAUUACCAUCACGC---AGUCAUUGAGCAAAUCAAUGGGGCUGCUCAUUAAUGU-------- .............((((((((......----...(((((....)))))......................((---(((((((((.....))))))..))))).)))))).))-------- ( -20.60, z-score = -0.34, R) >consensus GCCAAGAAAUUAUGCUUAAAGUUUUUUU__UAACGUGUAGCCAUACACAACCUAGUGAUAUUACCAUCACGC___AGUCAUUGAGCAAAUCAAUGGGGCUGCUCAUUAAUGU________ ..................................(.((((((............(((((......))))).......(((((((.....))))))))))))).)................ (-14.37 = -14.98 + 0.62)


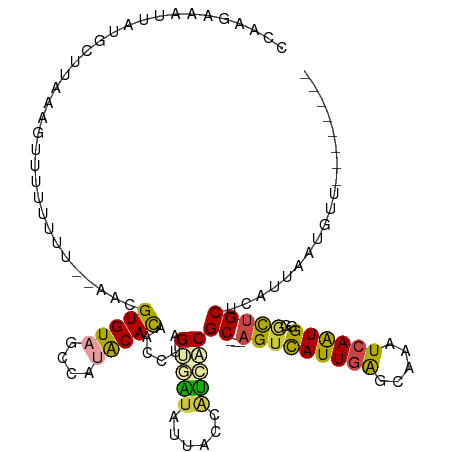
| Location | 22,861,964 – 22,862,073 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.61 |
| Shannon entropy | 0.45593 |
| G+C content | 0.37833 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -14.37 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22861964 109 + 27905053 CCAAAGUCUUACGCUUCAAGUUUUUUUUUUAACGUGUAGCCAUACACAACAUAGUGAUAUUACCAUCACGC---AGUCAUUGAGAAAAUCAAUGAGGCUGCUCAUUAAUGUU-------- ...................(((........)))(((((....)))))(((((.(((((......)))))((---(((((((((.....))))))..)))))......)))))-------- ( -24.50, z-score = -2.54, R) >droAna3.scaffold_12911 3777341 115 + 5364042 CCAAUAAACCAUUUAAAGAAUUUUCUCU-----GUGUAUCCACGCAUAACUGGGAGGUGUUCUCGCCUCGCCUGGGUUAUUCGGUUCAUCAAUGACUUGGCUCACUAAUGAGGUUUGCUU ....((((((.................(-----((((....)))))....(((((((((....))))))(((.((((((((.((....)))))))))))))...)))....))))))... ( -28.40, z-score = -0.95, R) >droEre2.scaffold_4820 5294957 106 - 10470090 CCAAAGUAUUAUGUUUAGAGAUUUGUUU--AACGUGUAGCCAAGCACAACAUAGUAAUAUUACCUUCUCGC---AGUCAUUGAGCAAGUCAAUG-GACUGCUCAUUAAUGUU-------- ......(((((((((((((......)))--)..((((......))))))))))))).............((---(((((((((.....))))).-))))))...........-------- ( -22.20, z-score = -1.15, R) >droYak2.chr3R 5200432 107 - 28832112 CCAAGGUAUUAUGCUUAAAGUUUUUUUU--AACGUGUAGCCAUGCACAACCUAGUGAUAUUACCAUCUCGC---AGUCAUUGAGCAAAUCAGUGGGGCUGCUCAUUAAUGUU-------- ....((((.((((((....(((......--)))(((((....))))).....))).))).)))).....((---(((((((((.....))))))..)))))...........-------- ( -25.00, z-score = -0.83, R) >droSec1.super_13 1625670 107 + 2104621 CCAAGAAAUUAUGGUAAAAGUUUUUUUU--AACGUGUAGCCAUACACAACCUAGUUGUAUUACCAUGACGC---AGUCAUUGAGCAAAUCAAUGGGGCUGCUCAUUAAUGUU-------- ....((..(((((((((..(((......--)))(((((....)))))............))))))))).((---(((((((((.....))))))..))))))).........-------- ( -27.70, z-score = -2.30, R) >droSim1.chr3R 22687640 105 + 27517382 CCAAGAAAUUAUGCUUAAUGUUUUUU----AACGUGUAGCCAUACACAACCUAGUUAUAUUACCAUCACGC---AGUCAUUGAGCAAAUCAAUGGGGCUGCUCAUUAAUGUU-------- ............((((((((......----...(((((....)))))......................((---(((((((((.....))))))..))))).)))))).)).-------- ( -20.70, z-score = -0.67, R) >consensus CCAAGAAAUUAUGCUUAAAGUUUUUUUU__AACGUGUAGCCAUACACAACCUAGUGAUAUUACCAUCACGC___AGUCAUUGAGCAAAUCAAUGGGGCUGCUCAUUAAUGUU________ .................................(.((((((............(((((......))))).......(((((((.....))))))))))))).)................. (-14.37 = -14.98 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:08 2011