| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,773,221 – 22,773,351 |
| Length | 130 |
| Max. P | 0.999388 |

| Location | 22,773,221 – 22,773,351 |
|---|---|
| Length | 130 |
| Sequences | 14 |
| Columns | 142 |
| Reading direction | forward |
| Mean pairwise identity | 67.14 |
| Shannon entropy | 0.71870 |
| G+C content | 0.42707 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -22.03 |
| Energy contribution | -21.66 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.84 |
| SVM RNA-class probability | 0.999388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
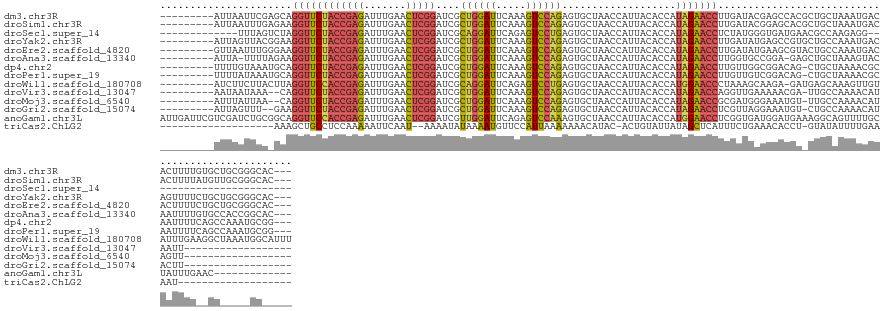
>dm3.chr3R 22773221 130 + 27905053 ---------AUUAAUUCGAGCAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUUGAUACGAGCCACGCUGCUAAAUGACACUUUUGUGCUGCGGGCAC--- ---------......(((..((((((((((((((.......)))))....((((((.....))))))...................))))))).))...)))(((.(((.((((((.......)))).)).))))))..--- ( -40.20, z-score = -3.10, R) >droSim1.chr3R 22604141 130 + 27517382 ---------AUUAAUUUGAGAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUUGAUACGGAGCACGCUGCUAAAUGACACUUUUAUGUUGCGGGCAC--- ---------...........((((((((((((((.......)))))....((((((.....))))))...................)))))))))........((.(((.((((((.......)))).)).))).))..--- ( -34.80, z-score = -2.02, R) >droSec1.super_14 1957689 105 + 2068291 -------------UUUAGUCUAGGUUCUACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUAGAACCUCUAUGGGUGAUGAACGCCAAGAGG------------------------ -------------.....(((.(((....(((((.......))))).....((.((((((....)))))).))...((((((.((((((.....))))))))))))...))).)))..------------------------ ( -34.70, z-score = -2.75, R) >droYak2.chr3R 5119663 130 - 28832112 ---------AUUAGUUACGGAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUUGAUAUGAGCCGUGCUGCCAAAUGACAGUUUUCUGCUGCGGGCAC--- ---------...(((.((((((((((((((((((.......)))))....((((((.....))))))...................)))))))))........)))))))(((....(.((((.....))))).)))..--- ( -38.10, z-score = -1.76, R) >droEre2.scaffold_4820 5204169 130 - 10470090 ---------GUUAAUUUGGGAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUUGAUAUGAAGCGUACUGCCAAAUGACACUUUUCUGCUGCGGGCAC--- ---------((((.(((((.((((((((((((((.......)))))....((((((.....))))))...................)))))))))..((((...))))...))))))))).......(((.....))).--- ( -34.20, z-score = -1.17, R) >droAna3.scaffold_13340 7143420 128 - 23697760 ---------AUUA-UUUUAGAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUUGGUGCCGGA-GAGCUGCUAAAGUACAAUUUUGUGCCACCGGCAC--- ---------....-......((((((((((((((.......)))))....((((((.....))))))...................))))))))).(((((((.-(.((.((.(((......))).))))).)))))))--- ( -41.80, z-score = -3.60, R) >dp4.chr2 26119616 129 - 30794189 ---------UUUUGUAAAUGCAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUUGUUGGCGGACAG-CUGCUAAAACGCAAUUUUCAGCCAAAUGCGG--- ---------.........((((((((((((((((.......)))))....((((((.....))))))...................)))))))...(((((.((.((-.(((......))).)).)).))))).)))).--- ( -38.30, z-score = -2.34, R) >droPer1.super_19 1496109 129 - 1869541 ---------UUUUAUAAAUGCAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUUGUUGUCGGACAG-CUGCUAAAACGCAAUUUUCAGCCAAAUGCGG--- ---------..........(((((((((((((((.......)))))....((((((.....))))))...................)))))))..((((.....)))-)))).....((((.(((......))))))).--- ( -32.60, z-score = -1.40, R) >droWil1.scaffold_180708 913136 132 + 12563649 ---------AUCUUCUUACUUAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCCUAAAGCAAGA-GAUGAGCAAAGUUGUAUUUGAAGGCUAAAUGGCAUUU ---------((((.(((.(((.((((((((((((.......))))).....((.((((((....)))))).)).............)))))))...))).))))-)))...((((......))))...(((....))).... ( -33.90, z-score = -0.98, R) >droVir3.scaffold_13047 6362780 112 - 19223366 ---------AAUAAUAAA--CAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCAGGUUGAAAAACGA-UUGCCAAAACAUAAUU------------------ ---------.........--..((((((((((((.......)))))....((((((.....))))))...................)))))))..(((....)))..-................------------------ ( -26.50, z-score = -2.83, R) >droMoj3.scaffold_6540 28994094 112 + 34148556 ---------AUUUAUUAA--CAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGCGAUGGGAAAUGU-UUGCCAAAACAUAGUU------------------ ---------.((((((..--..((((((((((((.......)))))....((((((.....))))))...................)))))))..))))))..((((-((....))))))....------------------ ( -28.40, z-score = -1.99, R) >droGri2.scaffold_15074 1846061 112 + 7742996 ---------AUUAGUUU--GAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUCGUUAGGAAAUGU-CUGCCAAAACAUACUU------------------ ---------....((((--((.((((((((((((.......)))))....((((((.....))))))...................)))))))))(.((((.....)-))).).))))......------------------ ( -26.90, z-score = -1.39, R) >anoGam1.chr3L 12834922 129 - 41284009 AUUGAUUCGUCGAUCUGCGGCAGGUUCCACCGAGAUUUGAACUCGGAUCGUUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUGGAACCUCGGUGAUGGAUGAAAGGCAGUUUUGCUAUUUGAAC------------- .....((((((.(((.((.(.(((((((((((((.......)))))....((((((.....))))))...................))))))))).))))).)))))).(((......)))........------------- ( -40.70, z-score = -1.79, R) >triCas2.ChLG2 12682797 100 + 12900155 -------------------AAAGCUGCCUCCAAAAAUUCAAU--AAAAUAUAAAAUGUUCCAUUAAAAAAACAUAC-ACUGUAUUAUAGCUCAUUUCUGAAACACCU-GUAUAUUUUGAAAAU------------------- -------------------......................(--((((((((...((((...........(((...-..)))........(((....)))))))...-.))))))))).....------------------- ( -5.70, z-score = 0.55, R) >consensus _________AUUAAUUAA_GAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUUGAUGCGAGACGA_CUGCCAAAUGACAAUUUU__GC_____GC_____ ......................((((((((((((.......)))))....((((((.....))))))...................)))))))................................................. (-22.03 = -21.66 + -0.37)
| Location | 22,773,221 – 22,773,351 |
|---|---|
| Length | 130 |
| Sequences | 14 |
| Columns | 142 |
| Reading direction | reverse |
| Mean pairwise identity | 67.14 |
| Shannon entropy | 0.71870 |
| G+C content | 0.42707 |
| Mean single sequence MFE | -31.69 |
| Consensus MFE | -22.87 |
| Energy contribution | -21.91 |
| Covariance contribution | -0.95 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.995736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
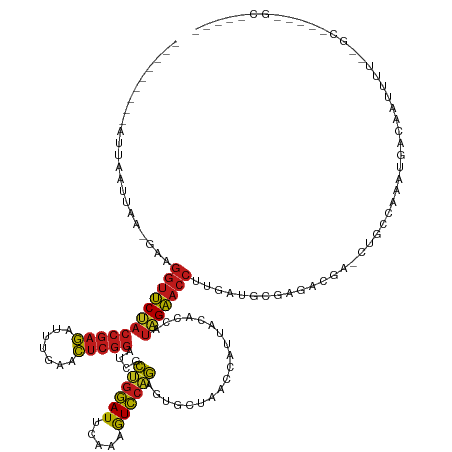
>dm3.chr3R 22773221 130 - 27905053 ---GUGCCCGCAGCACAAAAGUGUCAUUUAGCAGCGUGGCUCGUAUCAAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUGCUCGAAUUAAU--------- ---((((.....)))).........(((((((((.((.(((.(..(((((((((...(((((........)))))..)))))))))..).)))...(((((.......)))))....))))))).))))....--------- ( -37.60, z-score = -1.25, R) >droSim1.chr3R 22604141 130 - 27517382 ---GUGCCCGCAACAUAAAAGUGUCAUUUAGCAGCGUGCUCCGUAUCAAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUUCUCAAAUUAAU--------- ---..((.(((....((((.......))))...))).))........(((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))))...........--------- ( -30.20, z-score = -0.62, R) >droSec1.super_14 1957689 105 - 2068291 ------------------------CCUCUUGGCGUUCAUCACCCAUAGAGGUUCUAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUAGAACCUAGACUAAA------------- ------------------------((((((((.((.....))))).))))).....((((.((..((((....(.(((((.......))))).)..(((((.......)))))...)))))).))))..------------- ( -28.10, z-score = -0.79, R) >droYak2.chr3R 5119663 130 + 28832112 ---GUGCCCGCAGCAGAAAACUGUCAUUUGGCAGCACGGCUCAUAUCAAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUUCCGUAACUAAU--------- ---.((((....((((....)))).....))))..((((........(((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))))))).......--------- ( -36.30, z-score = -1.14, R) >droEre2.scaffold_4820 5204169 130 + 10470090 ---GUGCCCGCAGCAGAAAAGUGUCAUUUGGCAGUACGCUUCAUAUCAAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUUCCCAAAUUAAC--------- ---.(((.....)))..........((((((.....((((.....(((((((((...(((((........)))))..)))))))))....))))..(((((.......)))))..........))))))....--------- ( -32.00, z-score = -0.55, R) >droAna3.scaffold_13340 7143420 128 + 23697760 ---GUGCCGGUGGCACAAAAUUGUACUUUAGCAGCUC-UCCGGCACCAAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUUCUAAAA-UAAU--------- ---(((((((.(((.......(((......)))))).-.))))))).(((((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))))......-....--------- ( -40.61, z-score = -2.87, R) >dp4.chr2 26119616 129 + 30794189 ---CCGCAUUUGGCUGAAAAUUGCGUUUUAGCAG-CUGUCCGCCAACAAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUGCAUUUACAAAA--------- ---..((..(((((.((...((((......))))-...)).)))))..((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))))..........--------- ( -39.80, z-score = -2.32, R) >droPer1.super_19 1496109 129 + 1869541 ---CCGCAUUUGGCUGAAAAUUGCGUUUUAGCAG-CUGUCCGACAACAAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUGCAUUUAUAAAA--------- ---..((.....(((((((......)))))))..-.............((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))))..........--------- ( -33.50, z-score = -0.76, R) >droWil1.scaffold_180708 913136 132 - 12563649 AAAUGCCAUUUAGCCUUCAAAUACAACUUUGCUCAUC-UCUUGCUUUAGGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUAAGUAAGAAGAU--------- ..................................(((-((((((((..((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))))))))).))))--------- ( -36.00, z-score = -1.61, R) >droVir3.scaffold_13047 6362780 112 + 19223366 ------------------AAUUAUGUUUUGGCAA-UCGUUUUUCAACCUGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUG--UUUAUUAUU--------- ------------------......((((((...(-(((((.(((((...(((((...(((((........)))))..))))))))))...))))))(((((.......)))))))))))...--.........--------- ( -25.40, z-score = -0.92, R) >droMoj3.scaffold_6540 28994094 112 - 34148556 ------------------AACUAUGUUUUGGCAA-ACAUUUCCCAUCGCGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUG--UUAAUAAAU--------- ------------------......((..(((.((-....)).)))..))(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))..--.........--------- ( -26.10, z-score = -0.80, R) >droGri2.scaffold_15074 1846061 112 - 7742996 ------------------AAGUAUGUUUUGGCAG-ACAUUUCCUAACGAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUUC--AAACUAAU--------- ------------------.((((((((......)-))))........(((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))).--..)))...--------- ( -27.90, z-score = -0.95, R) >anoGam1.chr3L 12834922 129 + 41284009 -------------GUUCAAAUAGCAAAACUGCCUUUCAUCCAUCACCGAGGUUCCAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAACGAUCCGAGUUCAAAUCUCGGUGGAACCUGCCGCAGAUCGACGAAUCAAU -------------.........(((....)))..(((.((.(((..((((((((((((((((........)))))((((((((.((.((....)))).))))))))......)))))))))..))..))).)).)))..... ( -33.40, z-score = -0.90, R) >triCas2.ChLG2 12682797 100 - 12900155 -------------------AUUUUCAAAAUAUAC-AGGUGUUUCAGAAAUGAGCUAUAAUACAGU-GUAUGUUUUUUUAAUGGAACAUUUUAUAUUUU--AUUGAAUUUUUGGAGGCAGCUUU------------------- -------------------.((((((((((((((-(.((((((((....))).....)))))..)-))))))...((((((((((.........))))--))))))..)))))))).......------------------- ( -16.80, z-score = -0.80, R) >consensus _____GC_____GC__AAAAUUGUCAUUUAGCAG_ACGUCUCCCAUCAAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUGC_UUAAUUAAU_________ ................................................((((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))))..................... (-22.87 = -21.91 + -0.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:03 2011