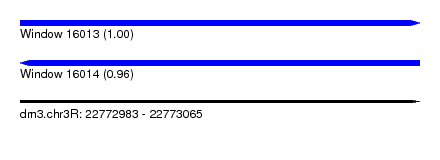
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,772,983 – 22,773,065 |
| Length | 82 |
| Max. P | 0.995595 |

| Location | 22,772,983 – 22,773,065 |
|---|---|
| Length | 82 |
| Sequences | 12 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 82.90 |
| Shannon entropy | 0.40671 |
| G+C content | 0.45173 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -20.16 |
| Energy contribution | -20.49 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
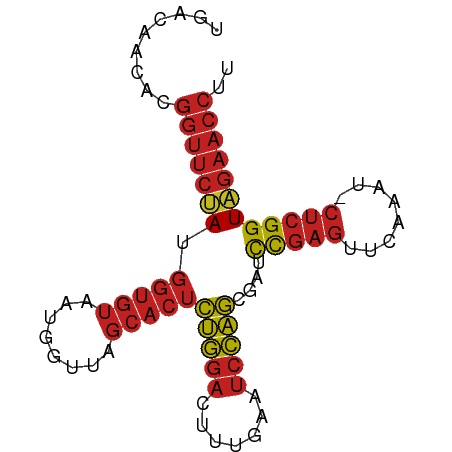
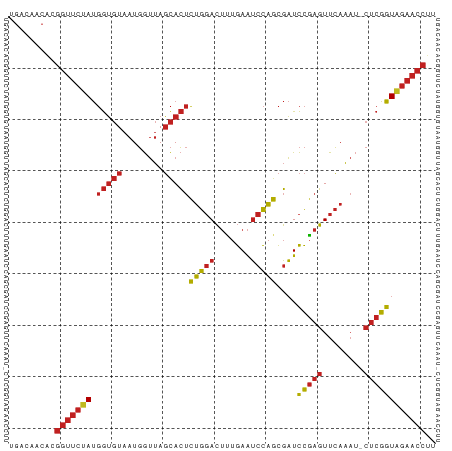
>dm3.chr3R 22772983 82 + 27905053 AAGGUUCUACCGAG-AUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGCGUUGGCA ..((((((((((((-.......)))))....((((((.....))))))...................)))))))((....)). ( -25.80, z-score = -2.76, R) >droSim1.chr3R 22603902 82 + 27517382 AAGGUUCUACCGAG-AUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGCGAUGUCA ..((((((((((((-.......)))))....((((((.....))))))...................)))))))......... ( -24.40, z-score = -2.65, R) >droSec1.super_14 1957473 79 + 2068291 CAGGUUCCACCGAG-AUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUCGCUG--- .(((((((((((((-.......))))).....((.((((((....)))))).)).............)))))))).....--- ( -27.00, z-score = -2.97, R) >droYak2.chr3R 5119425 82 - 28832112 AAGGUUCUACCGAG-AUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGCGUUGUCA ..((((((((((((-.......)))))....((((((.....))))))...................)))))))......... ( -24.40, z-score = -2.56, R) >droEre2.scaffold_4820 5203929 82 - 10470090 AAGGUUCUACCGAG-AUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGCGUCUUCA ..((((((((((((-.......)))))....((((((.....))))))...................)))))))......... ( -24.40, z-score = -2.77, R) >droAna3.scaffold_13340 7143042 82 - 23697760 CAGGUUCUACCGAG-AUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGUGUUGUCU ..((((((((((((-.......)))))....((((((.....))))))...................)))))))......... ( -24.40, z-score = -2.40, R) >dp4.chr2 26119366 82 - 30794189 AAGGUUCUACCGAG-AUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGUGUUGUCA ..((((((((((((-.......)))))....((((((.....))))))...................)))))))......... ( -24.40, z-score = -2.43, R) >droPer1.super_19 1495859 82 - 1869541 AAGGUUCUACCGAG-AUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGUGCUGUCA ..((((((((((((-.......)))))....((((((.....))))))...................)))))))......... ( -24.40, z-score = -2.41, R) >droWil1.scaffold_181130 6044058 82 + 16660200 AAGGUUCUACCGAG-AUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCUUGUUGACA ((((((((((((((-.......)))))....((((((.....))))))...................)))))))))....... ( -25.70, z-score = -3.32, R) >droGri2.scaffold_15074 1845805 82 + 7742996 CAGGUUCUACCGAG-AUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGUGCUGACA ..((((((((((((-.......)))))....((((((.....))))))...................)))))))......... ( -24.40, z-score = -2.58, R) >anoGam1.chr3L 12831886 82 - 41284009 UAGGUUCCACCGAG-AUUUGAACUCGGAUCGUUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUGGAACCCGGUUGACA ..((((((((((((-.......)))))....((((((.....))))))...................)))))))......... ( -23.50, z-score = -1.49, R) >triCas2.ChLG2 12682557 71 + 12900155 --------ACCGAAUGGCUCAAAGAAAAAAAUUAGUCUUAUUUUGUAAAUUAAUGUUAAUUAAUUGGUUCACUUUGGUC---- --------(((((((((..(.((((..........))))........(((((((....))))))))..))).)))))).---- ( -8.80, z-score = -0.26, R) >consensus AAGGUUCUACCGAG_AUUUGAACUCGGAUCGCUGGAUUCAAAGUCCAGAGUGCUAACCAUUACACCAUAGAACCGCGUUGUCA ..((((((((((((........)))))....((((((.....))))))...................)))))))......... (-20.16 = -20.49 + 0.33)

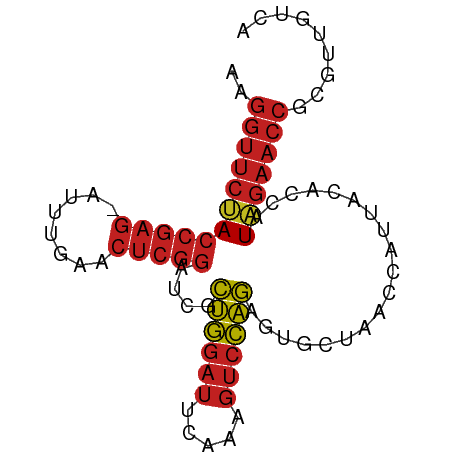
| Location | 22,772,983 – 22,773,065 |
|---|---|
| Length | 82 |
| Sequences | 12 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 82.90 |
| Shannon entropy | 0.40671 |
| G+C content | 0.45173 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -19.45 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22772983 82 - 27905053 UGCCAACGCGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAU-CUCGGUAGAACCUU .((....))(((((((.(((((........)))))(((((.......)))))....(((((.......-)))))))))))).. ( -24.10, z-score = -1.62, R) >droSim1.chr3R 22603902 82 - 27517382 UGACAUCGCGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAU-CUCGGUAGAACCUU .........(((((((.(((((........)))))(((((.......)))))....(((((.......-)))))))))))).. ( -23.70, z-score = -1.48, R) >droSec1.super_14 1957473 79 - 2068291 ---CAGCGAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAU-CUCGGUGGAACCUG ---.....((((((((.(((((........)))))(((((.......)))))....(((((.......-))))))))))))). ( -26.90, z-score = -1.93, R) >droYak2.chr3R 5119425 82 + 28832112 UGACAACGCGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAU-CUCGGUAGAACCUU .........(((((((.(((((........)))))(((((.......)))))....(((((.......-)))))))))))).. ( -23.70, z-score = -1.57, R) >droEre2.scaffold_4820 5203929 82 + 10470090 UGAAGACGCGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAU-CUCGGUAGAACCUU .........(((((((.(((((........)))))(((((.......)))))....(((((.......-)))))))))))).. ( -23.70, z-score = -1.37, R) >droAna3.scaffold_13340 7143042 82 + 23697760 AGACAACACGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAU-CUCGGUAGAACCUG .........(((((((.(((((........)))))(((((.......)))))....(((((.......-)))))))))))).. ( -23.70, z-score = -1.65, R) >dp4.chr2 26119366 82 + 30794189 UGACAACACGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAU-CUCGGUAGAACCUU .........(((((((.(((((........)))))(((((.......)))))....(((((.......-)))))))))))).. ( -23.70, z-score = -1.72, R) >droPer1.super_19 1495859 82 + 1869541 UGACAGCACGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAU-CUCGGUAGAACCUU .........(((((((.(((((........)))))(((((.......)))))....(((((.......-)))))))))))).. ( -23.70, z-score = -1.46, R) >droWil1.scaffold_181130 6044058 82 - 16660200 UGUCAACAAGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAU-CUCGGUAGAACCUU .......(((((((((.(((((........)))))(((((.......)))))....(((((.......-)))))))))))))) ( -25.00, z-score = -2.35, R) >droGri2.scaffold_15074 1845805 82 - 7742996 UGUCAGCACGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAU-CUCGGUAGAACCUG .........(((((((.(((((........)))))(((((.......)))))....(((((.......-)))))))))))).. ( -23.70, z-score = -1.45, R) >anoGam1.chr3L 12831886 82 + 41284009 UGUCAACCGGGUUCCAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAACGAUCCGAGUUCAAAU-CUCGGUGGAACCUA ........((((((((((((((........)))))((((((((.((.((....)))).))))))))..-....))))))))). ( -27.80, z-score = -2.32, R) >triCas2.ChLG2 12682557 71 - 12900155 ----GACCAAAGUGAACCAAUUAAUUAACAUUAAUUUACAAAAUAAGACUAAUUUUUUUCUUUGAGCCAUUCGGU-------- ----.(((..((((....(((((((....)))))))..((((..((((......))))..))))...)))).)))-------- ( -5.10, z-score = 0.38, R) >consensus UGACAACACGGUUCUAUGGUGUAAUGGUUAGCACUCUGGACUUUGAAUCCAGCGAUCCGAGUUCAAAU_CUCGGUAGAACCUU .........(((((((.(((((........)))))(((((.......)))))....(((((........)))))))))))).. (-19.45 = -19.80 + 0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:01 2011