| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,757,311 – 22,757,401 |
| Length | 90 |
| Max. P | 0.951181 |

| Location | 22,757,311 – 22,757,401 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 72.13 |
| Shannon entropy | 0.57859 |
| G+C content | 0.43493 |
| Mean single sequence MFE | -21.61 |
| Consensus MFE | -11.23 |
| Energy contribution | -11.14 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
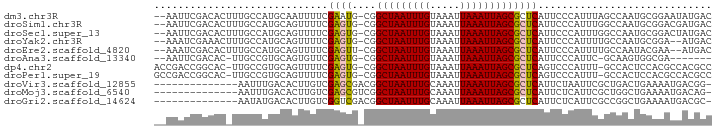
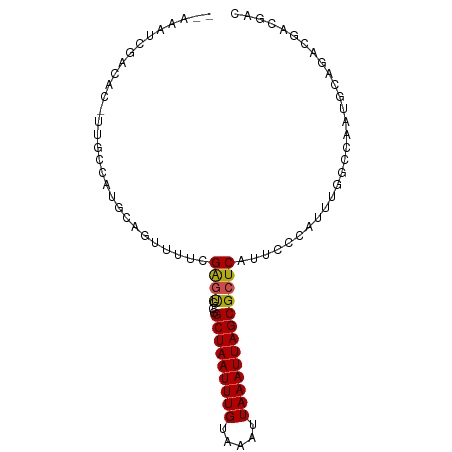
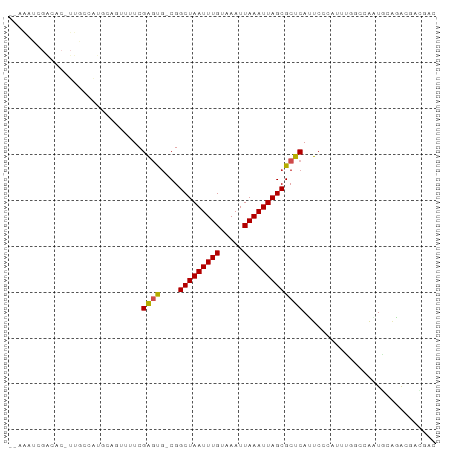
>dm3.chr3R 22757311 90 - 27905053 --AAUUCGACACUUUGCCAUGCAAUUUUCGAAUG-CGGCUAAUUUGUAAAUUAAAUUAGCGCUCAUUCCCAUUUAGCCAAUGCGGAAUAUGAC --.((((((....((((...))))...))))))(-((.((((((((.....)))))))))))..(((((((((.....)))).)))))..... ( -23.60, z-score = -2.81, R) >droSim1.chr3R 22587717 90 - 27517382 --AAUUCGACACUUUGCCAUGCAGUUUUCGAGUG-CGGCUAAUUUGUAAAUUAAAUUAGCGCUCAUUCCCAUUUGGCCAAUGCGGACGAUGAC --.((((((....((((...))))...))))))(-((.((((((((.....)))))))))))(((((((((((.....)))).)))..)))). ( -22.70, z-score = -1.10, R) >droSec1.super_13 1529920 90 - 2104621 --AAUUCGACACUUUGCCAUGCAGUUUUCGAGUG-CGGCUAAUUUGUAAAUUAAAUUAGCGCUCAUUCCCAUUUGGCCAAUGCGGACUAUGAC --.((((((....((((...))))...))))))(-((.((((((((.....)))))))))))(((((((((((.....)))).)))..)))). ( -22.70, z-score = -1.43, R) >droYak2.chr3R 5103362 88 + 28832112 --AAAUCGAAACUUUGCCAUGCAGUUUUCGAGUG-CGGCUAAUUUGUAAAUUAAAUUAGCGCUCAUUCCCAUUUUGCCAAUGCGGA--AUGAC --...((((((..((((...)))).))))))..(-((.((((((((.....)))))))))))(((((((((((.....)))).)))--)))). ( -27.90, z-score = -3.64, R) >droEre2.scaffold_4820 5187729 88 + 10470090 --AAAUCGACACUUUGCCAUGCAGUUUUCGAGUU-CGGCUAAUUUGUAAAUUAAAUUAGCGCUCAUUCCCAUUUUGCCAAUACGAA--AUGAC --.....((....((((...))))...))((((.-..(((((((((.....))))))))))))).....(((((((......))))--))).. ( -18.60, z-score = -1.95, R) >droAna3.scaffold_13340 7126525 81 + 23697760 --AAUUCGACAC-UUGCCGUGCAGUGUUCGAGUG-CGGCUAAUUUGUAAAUUAAAUUAGCGCUCAUUCCCAUUC-GCAAGUGGCGA------- --...(((.(((-((((...(.((((...(((((-.((((((((((.....))))))))).).))))).)))))-))))))).)))------- ( -26.80, z-score = -2.63, R) >dp4.chr2 26101911 90 + 30794189 ACCGACCGGCAC-UUGCCGUGCAGUUUUCGAGUG-CGGCUAAUUUGUAAAUUAAAUUAGCGCUCAGUCCCAUUU-GCCACUCCACGCCACGCC ...(..((((..-..))))..).......(((((-..(((((((((.....)))))))))((............-)))))))........... ( -22.00, z-score = -0.68, R) >droPer1.super_19 1478023 90 + 1869541 GCCGACCGGCAC-UUGCCGUGCAGUUUUCGAGUG-CGGCUAAUUUGUAAAUUAAAUUAGCGCUCAGUCCCAUUU-GCCACUCCACGCCACGCC (((....)))..-....((((..((....(((((-..(((((((((.....)))))))))((............-))))))).))..)))).. ( -22.80, z-score = -0.57, R) >droVir3.scaffold_12855 2565684 78 - 10161210 --------------AAUUUGACACUUGUCGAGCGACGGCUAAUUUGCAAAUUAAAUUAGCGCUCAUUCUAAUUCGCUGACUGAAAAUGACGG- --------------...........((((((((....(((((((((.....))))))))))))).......((((.....))))...)))).- ( -16.00, z-score = -1.05, R) >droMoj3.scaffold_6540 11771140 78 - 34148556 --------------AAUUUGACACUUGUCGAGCGUCGGCUAAUUUGCAAAUUAAAUUAGCGCUCAUUCUCAUUCGCUGGCUGAAAAUGACAG- --------------.....(((....)))((((....(((((((((.....)))))))))))))....(((((..(.....)..)))))...- ( -17.70, z-score = -0.89, R) >droGri2.scaffold_14624 1861121 78 + 4233967 --------------AAUAUGACACUUGUCGGUCGACGGCUAAUUUGCAAAUUAAAUUAGCGCUCAUUCUCAUUCGCCGGCUGAAAAUGACGC- --------------.......((....(((((((...(((((((((.....)))))))))((............)))))))))...))....- ( -16.90, z-score = -0.55, R) >consensus __AAAUCGACAC_UUGCCAUGCAGUUUUCGAGUG_CGGCUAAUUUGUAAAUUAAAUUAGCGCUCAUUCCCAUUUGGCCAAUGCAGACGACGAC .............................((((....(((((((((.....)))))))))))))............................. (-11.23 = -11.14 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:00 2011