
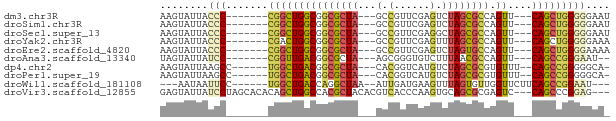
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,736,228 – 22,736,290 |
| Length | 62 |
| Max. P | 0.999825 |

| Location | 22,736,228 – 22,736,290 |
|---|---|
| Length | 62 |
| Sequences | 10 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 69.59 |
| Shannon entropy | 0.58600 |
| G+C content | 0.57426 |
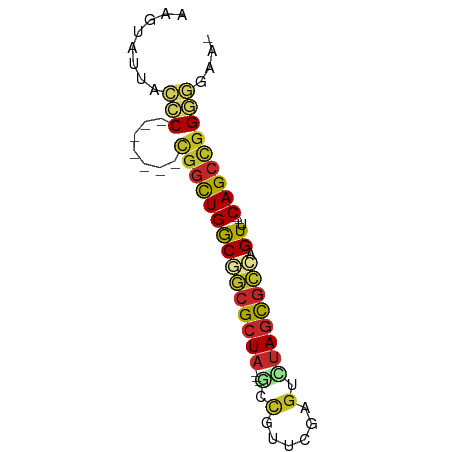
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -13.69 |
| Energy contribution | -13.50 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.77 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
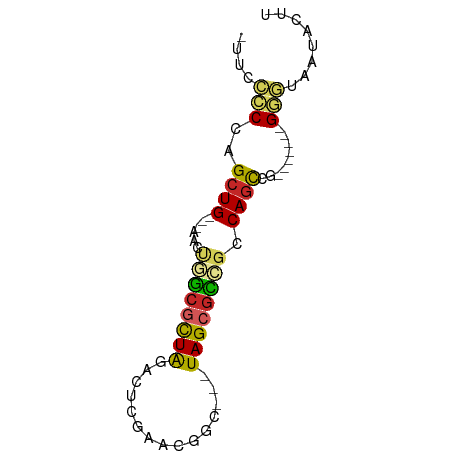
>dm3.chr3R 22736228 62 + 27905053 AAGUAUUACCC-------CGGCUGGCGGCGCUA---GCCGUUCGAGUCUAGCGCCAGUU---CAGCUGGGGGAAU ........(((-------(((((((((((((((---(.(......).)))))))).).)---))))))))).... ( -36.00, z-score = -4.83, R) >droSim1.chr3R 22567031 62 + 27517382 AAGUAUUACCC-------CGGCUGGCGGCGCUA---GCCGUUCGAGUCUAGCGCCAGUU---CAGCUGGGGGAAU ........(((-------(((((((((((((((---(.(......).)))))))).).)---))))))))).... ( -36.00, z-score = -4.83, R) >droSec1.super_13 1509244 62 + 2104621 AAGUAUUACCC-------CGGCUGGCGGCGCUA---GCCGUUCGAGGCUAGCGCCAGUU---CAGCUGGGGGAAU ........(((-------(((((((((((((((---(((......)))))))))).).)---))))))))).... ( -43.10, z-score = -6.59, R) >droYak2.chr3R 5080742 62 - 28832112 AAGUAUUACCC-------CGACUGGCGGCGCUA---GCCGUUCGAGUUUAGCGCCAGUU---CAGCUGGGGGAAA ........(((-------((.((((((((((((---(.(......).)))))))).).)---))).))))).... ( -27.20, z-score = -2.51, R) >droEre2.scaffold_4820 5166601 62 - 10470090 AAGUAUUACCC-------CGGCUGGCGGCGCUA---GCCGUUCGAGUCUAGUGCCAGUU---CAGCUGGGGAAAA ........(((-------(((((((((((((((---(.(......).)))))))).).)---))))))))).... ( -33.90, z-score = -4.80, R) >droAna3.scaffold_13340 7103758 60 - 23697760 UAGUAUUAUCC-------CGGUUGACGGCGCUA---AGCGGGUGUCUUUAACGCCAGUU---CAGCCGGGAAU-- ........(((-------((((((((.((....---.)).(((((.....))))).).)---)))))))))..-- ( -26.00, z-score = -3.50, R) >dp4.chr2 26077117 63 - 30794189 AAGUAUUAAGCC------UGGCUGACGGCGCUA---CACGGUCAUGUCUAGCGCGUGUUU--CAGCCGGGGGCA- ..........((------(((((((((((((((---.(((....))).)))))).))..)--))))))))....- ( -27.80, z-score = -2.54, R) >droPer1.super_19 1453604 63 - 1869541 AAGUAUUAAGCC------UGGCUGACGGCGCUA---CACGGUCAUGUCUAGCGCGUGUUU--CAGCCGGGGGCA- ..........((------(((((((((((((((---.(((....))).)))))).))..)--))))))))....- ( -27.80, z-score = -2.54, R) >droWil1.scaffold_181108 514015 61 + 4707319 ---AAUAAUUCC------UGGCUGACCAGGCUAA--AUUGAUGAAGUUUAGUGUUGGUUCUUCAGCCGGAAU--- ---....(((((------.(((((((((((((((--(((.....)))))))).))))....)))))))))))--- ( -23.50, z-score = -4.45, R) >droVir3.scaffold_12855 2534048 69 + 10161210 GAGUAUUAUCCUAGCACACAGCUGGCCACGCUACACGUCACCCAAGUGCAGCGCGAGUC---CAGCCCGGAG--- ........(((..(....).(((((((.((((.(((.........))).)))).).).)---))))..))).--- ( -18.90, z-score = -1.13, R) >consensus AAGUAUUACCC_______CGGCUGGCGGCGCUA___GCCGUUCGAGUCUAGCGCCAGUU___CAGCCGGGGGAA_ ........(((.......(((((((((((((((...............))))))).))....))))))))).... (-13.69 = -13.50 + -0.19)
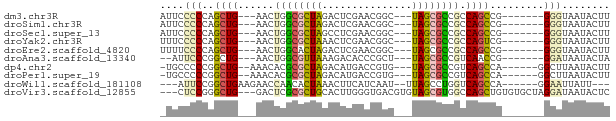
| Location | 22,736,228 – 22,736,290 |
|---|---|
| Length | 62 |
| Sequences | 10 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 69.59 |
| Shannon entropy | 0.58600 |
| G+C content | 0.57426 |
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -10.25 |
| Energy contribution | -9.83 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.79 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.00 |
| SVM RNA-class probability | 0.999549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22736228 62 - 27905053 AUUCCCCCAGCUG---AACUGGCGCUAGACUCGAACGGC---UAGCGCCGCCAGCCG-------GGGUAAUACUU (((.((((.((((---...(((((((((.((.....)))---)))))))).)))).)-------))).))).... ( -30.40, z-score = -4.12, R) >droSim1.chr3R 22567031 62 - 27517382 AUUCCCCCAGCUG---AACUGGCGCUAGACUCGAACGGC---UAGCGCCGCCAGCCG-------GGGUAAUACUU (((.((((.((((---...(((((((((.((.....)))---)))))))).)))).)-------))).))).... ( -30.40, z-score = -4.12, R) >droSec1.super_13 1509244 62 - 2104621 AUUCCCCCAGCUG---AACUGGCGCUAGCCUCGAACGGC---UAGCGCCGCCAGCCG-------GGGUAAUACUU (((.((((.((((---...(((((((((((......)))---)))))))).)))).)-------))).))).... ( -36.90, z-score = -5.90, R) >droYak2.chr3R 5080742 62 + 28832112 UUUCCCCCAGCUG---AACUGGCGCUAAACUCGAACGGC---UAGCGCCGCCAGUCG-------GGGUAAUACUU ....((((.((((---...((((((((..(......)..---)))))))).)))).)-------)))........ ( -26.30, z-score = -3.16, R) >droEre2.scaffold_4820 5166601 62 + 10470090 UUUUCCCCAGCUG---AACUGGCACUAGACUCGAACGGC---UAGCGCCGCCAGCCG-------GGGUAAUACUU ....((((.((((---...((((.((((.((.....)))---))).)))).)))).)-------)))........ ( -24.60, z-score = -2.76, R) >droAna3.scaffold_13340 7103758 60 + 23697760 --AUUCCCGGCUG---AACUGGCGUUAAAGACACCCGCU---UAGCGCCGUCAACCG-------GGAUAAUACUA --..((((((.((---(...((((((((.(.....)..)---))))))).))).)))-------)))........ ( -25.00, z-score = -4.28, R) >dp4.chr2 26077117 63 + 30794189 -UGCCCCCGGCUG--AAACACGCGCUAGACAUGACCGUG---UAGCGCCGUCAGCCA------GGCUUAAUACUU -.(((...(((((--(.....((((((.((......)).---))))))..)))))).------)))......... ( -26.50, z-score = -4.13, R) >droPer1.super_19 1453604 63 + 1869541 -UGCCCCCGGCUG--AAACACGCGCUAGACAUGACCGUG---UAGCGCCGUCAGCCA------GGCUUAAUACUU -.(((...(((((--(.....((((((.((......)).---))))))..)))))).------)))......... ( -26.50, z-score = -4.13, R) >droWil1.scaffold_181108 514015 61 - 4707319 ---AUUCCGGCUGAAGAACCAACACUAAACUUCAUCAAU--UUAGCCUGGUCAGCCA------GGAAUUAUU--- ---(((((((((((....(((...(((((.........)--))))..))))))))).------)))))....--- ( -16.40, z-score = -3.23, R) >droVir3.scaffold_12855 2534048 69 - 10161210 ---CUCCGGGCUG---GACUCGCGCUGCACUUGGGUGACGUGUAGCGUGGCCAGCUGUGUGCUAGGAUAAUACUC ---.(((.(((((---(.(.((((((((((...(....))))))))))))))))))(....)..)))........ ( -29.90, z-score = -2.20, R) >consensus _UUCCCCCAGCUG___AACUGGCGCUAGACUCGAACGGC___UAGCGCCGCCAGCCG_______GGGUAAUACUU .........((((......((((((((...((....))....)))))))).)))).................... (-10.25 = -9.83 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:58 2011