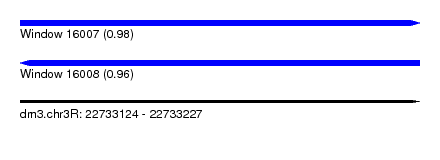
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,733,124 – 22,733,227 |
| Length | 103 |
| Max. P | 0.979836 |

| Location | 22,733,124 – 22,733,227 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 69.97 |
| Shannon entropy | 0.52737 |
| G+C content | 0.42850 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -17.12 |
| Energy contribution | -18.48 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
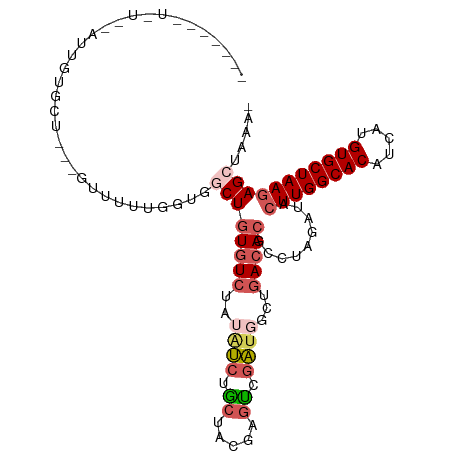
>dm3.chr3R 22733124 103 + 27905053 --AUGUUUUUUAUUUGUGCU---GUUUUUGGAGGCUGUGUCUAUAUCUACUGCGAGUAGAUGGCUGACACGCCUAGAUACUUGGCACGUCAUGUGCUAAGAGCUCAAA --..........((((.(((---........((((.(((((..(((((((.....)))))))...))))))))).....((((((((.....))))))))))).)))) ( -38.30, z-score = -4.05, R) >droAna3.scaffold_13340 7100758 90 - 23697760 ------------AUUUUUCUC--CUCUUUUGUAUCUAUAUAUGUGCCUA-UAGCUGUGGGU--CUGACACGCCUAGACACUUGGCACCACAUGUGCUAAGAGCCUCA- ------------.........--(((....((.((((....((((((((-(....))))))--...)))....)))).))(((((((.....)))))))))).....- ( -18.40, z-score = 0.15, R) >droSim1.chr3R 22563969 86 + 27517382 ------------------AU---GUUUUUUUGUGCUGUGUCUAUAUCUGCUACGAGUCGAUGGCUGACACGCCUAGAUACUUGGCACAUCAUGUGCUAAGAGCUAAA- ------------------..---..........((((((((..((((.((.....)).))))...))))).........((((((((.....)))))))))))....- ( -25.70, z-score = -1.90, R) >droSec1.super_13 1506227 98 + 2104621 ------AUGUUUUUUGUGCU---GUAUUUGGUGGCUGUGUCUAUAUCUGCUACGAGUCGAUGGCUGACACGCCUAGAUACUUGGCACAUCAUGUGCUAAGAGCUAAA- ------...........(((---.((((((..(((.(((((..((((.((.....)).))))...))))))))))))))((((((((.....)))))))))))....- ( -31.50, z-score = -2.16, R) >droYak2.chr3R 5077577 107 - 28832112 AUGUUUUUUUGUAUUGUGUUUGUGUUUUUGGUGCCUGUGUCUAUAUCUGCUGACUGCUAACUGCUGACACGCCUAGAUAGUUGGCACAUCAUGUGCUAAGAGCUAAA- .......................((((((((..(.((((((..(((((((((.(.((.....)).).)).))..)))))...))))))....)..))))))))....- ( -26.30, z-score = -0.76, R) >consensus _______U_U__AUUGUGCU___GUUUUUGGUGGCUGUGUCUAUAUCUGCUACGAGUCGAUGGCUGACACGCCUAGAUACUUGGCACAUCAUGUGCUAAGAGCUAAA_ .................................((((((((..((((.((.....)).))))...))))).........((((((((.....)))))))))))..... (-17.12 = -18.48 + 1.36)

| Location | 22,733,124 – 22,733,227 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.97 |
| Shannon entropy | 0.52737 |
| G+C content | 0.42850 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -12.48 |
| Energy contribution | -12.60 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
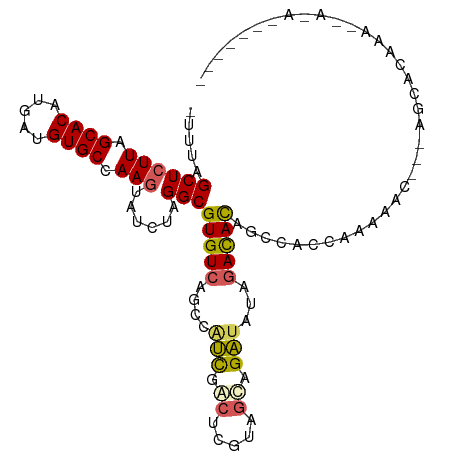
>dm3.chr3R 22733124 103 - 27905053 UUUGAGCUCUUAGCACAUGACGUGCCAAGUAUCUAGGCGUGUCAGCCAUCUACUCGCAGUAGAUAUAGACACAGCCUCCAAAAAC---AGCACAAAUAAAAAACAU-- ((((.((((((.((((.....)))).))).....(((((((((....((((((.....))))))...))))).))))........---))).))))..........-- ( -28.50, z-score = -3.91, R) >droAna3.scaffold_13340 7100758 90 + 23697760 -UGAGGCUCUUAGCACAUGUGGUGCCAAGUGUCUAGGCGUGUCAG--ACCCACAGCUA-UAGGCACAUAUAUAGAUACAAAAGAG--GAGAAAAAU------------ -.....(((((.((((.....))))...(((((((((((((....--...))).))).-)))))))..............)))))--.........------------ ( -23.30, z-score = -1.25, R) >droSim1.chr3R 22563969 86 - 27517382 -UUUAGCUCUUAGCACAUGAUGUGCCAAGUAUCUAGGCGUGUCAGCCAUCGACUCGUAGCAGAUAUAGACACAGCACAAAAAAAC---AU------------------ -....(((....((((.....))))...((((((..(((.(((.......))).)))...))))))......)))..........---..------------------ ( -19.20, z-score = -1.13, R) >droSec1.super_13 1506227 98 - 2104621 -UUUAGCUCUUAGCACAUGAUGUGCCAAGUAUCUAGGCGUGUCAGCCAUCGACUCGUAGCAGAUAUAGACACAGCCACCAAAUAC---AGCACAAAAAACAU------ -....((((((.((((.....)))).)))......((((((((....(((..(.....)..)))...))))).))).........---)))...........------ ( -21.90, z-score = -1.71, R) >droYak2.chr3R 5077577 107 + 28832112 -UUUAGCUCUUAGCACAUGAUGUGCCAACUAUCUAGGCGUGUCAGCAGUUAGCAGUCAGCAGAUAUAGACACAGGCACCAAAAACACAAACACAAUACAAAAAAACAU -....((.....)).......(((((..((....))..(((((.((.....)).(((....)))...))))).))))).............................. ( -18.90, z-score = -0.65, R) >consensus _UUUAGCUCUUAGCACAUGAUGUGCCAAGUAUCUAGGCGUGUCAGCCAUCGACUCGUAGCAGAUAUAGACACAGCCACCAAAAAC___AGCACAAA__A_A_______ .....((((((.((((.....)))).)))......)))(((((....(((.((.....)).)))...))))).................................... (-12.48 = -12.60 + 0.12)

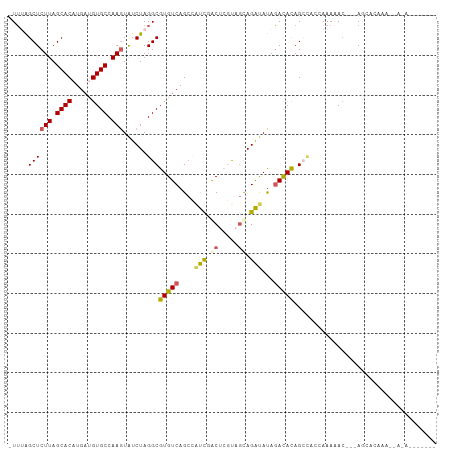
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:57 2011