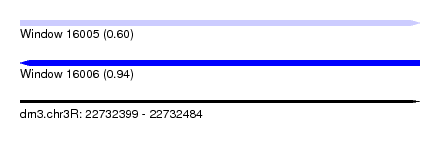
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,732,399 – 22,732,484 |
| Length | 85 |
| Max. P | 0.935174 |

| Location | 22,732,399 – 22,732,484 |
|---|---|
| Length | 85 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 70.86 |
| Shannon entropy | 0.52931 |
| G+C content | 0.33797 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -7.91 |
| Energy contribution | -8.74 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598918 |
| Prediction | RNA |
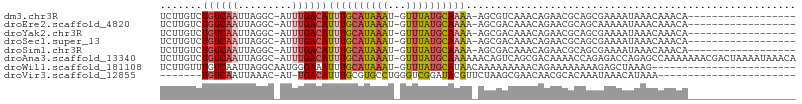
Download alignment: ClustalW | MAF
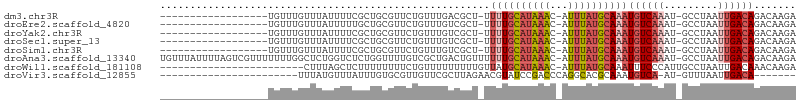
>dm3.chr3R 22732399 85 + 27905053 ------------------UGUUUGUUUAUUUUCGCUGCGUUCUGUUUGACGCU-UUUUGCAUAAAC-AUUUAUGCAAAUGUCAAAU-GCCUAAUUGACAGACAAGA ------------------((((((((.(((...((.(((((......))))).-.(((((((((..-..)))))))))........-))..))).))))))))... ( -22.90, z-score = -2.79, R) >droEre2.scaffold_4820 5162647 85 - 10470090 ------------------UGUUUGUUUAUUUUUGCUGCGUUCUGUUUGUCGCU-UUUUGCAUAAAC-AUUUAUGCAAAUGUCAAAU-GCCUAAUUGACAGACAAGA ------------------..(((((((........(((((..(((((((.((.-....))))))))-)...)))))...(((((..-......)))))))))))). ( -20.40, z-score = -1.88, R) >droYak2.chr3R 5076822 85 - 28832112 ------------------UGUUUGUUUAUUUUCGCUGCGUUCUGUUUGUCGCU-UUUUGCAUAAAC-AUUUAUGCAAAUGUCAAAU-GCCUAAUUGACAGACAAGA ------------------..(((((((........(((((..(((((((.((.-....))))))))-)...)))))...(((((..-......)))))))))))). ( -20.40, z-score = -1.90, R) >droSec1.super_13 1505512 85 + 2104621 ------------------UGUUUGUUUAUUUUCGCUGCGUUCUGUUUGUCGCU-UUUUGCAUAAAC-AUUUAUGCAAAUGUCAAAU-GCCUAAUUGACAGACAAGA ------------------..(((((((........(((((..(((((((.((.-....))))))))-)...)))))...(((((..-......)))))))))))). ( -20.40, z-score = -1.90, R) >droSim1.chr3R 22563249 85 + 27517382 ------------------UGUUUGUUUAUUUUCGCUGCGUUCUGUUUGUCGCU-UUUUGCAUAAAC-AUUUAUGCAAAUGUCAAAU-GCCUAAUUGACAGACAAGA ------------------..(((((((........(((((..(((((((.((.-....))))))))-)...)))))...(((((..-......)))))))))))). ( -20.40, z-score = -1.90, R) >droAna3.scaffold_13340 7100239 104 - 23697760 UGUUUAUUUUAGUCGUUUUUUUGGCUCUGGUCUCUGGUUUUGUCGCUGACUGUUUUUUGCAUAAAC-AUUUAUGCAAAUGUCAAAU-GCCUAAUUGACAGACAAGA ...........((((((..((.(((...((((..(((.....)))..))))(...(((((((((..-..)))))))))...)....-))).))..))).))).... ( -22.60, z-score = -1.13, R) >droWil1.scaffold_181108 510141 81 + 4707319 ------------------------CUUUAGCUCUUUUUUUUCUGUUUUUUUUUGUUAUGCAUAAAC-AUUUAUGCAAAUUUCCCAUUGCCUAAUUGACAAACAAGA ------------------------.........................(((((((.(((((((..-..)))))))..........((.(.....).))))))))) ( -8.40, z-score = -0.18, R) >droVir3.scaffold_12855 2528361 74 + 10161210 -----------------------UUUAUGUUUAUUUGUGCGUUGUUCGCUUAGAACGUAUCCGACCCAGGCACGCAAAUGUCA-AU-GUUUAAUUGACA------- -----------------------.........(((((((.(((((((.....))))((.....))...))).)))))))((((-((-.....)))))).------- ( -16.90, z-score = -1.14, R) >consensus __________________UGUUUGUUUAUUUUCGCUGCGUUCUGUUUGUCGCU_UUUUGCAUAAAC_AUUUAUGCAAAUGUCAAAU_GCCUAAUUGACAGACAAGA .......................................................((((((((((...))))))))))((((((.........))))))....... ( -7.91 = -8.74 + 0.83)
| Location | 22,732,399 – 22,732,484 |
|---|---|
| Length | 85 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 70.86 |
| Shannon entropy | 0.52931 |
| G+C content | 0.33797 |
| Mean single sequence MFE | -17.51 |
| Consensus MFE | -9.35 |
| Energy contribution | -9.71 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935174 |
| Prediction | RNA |
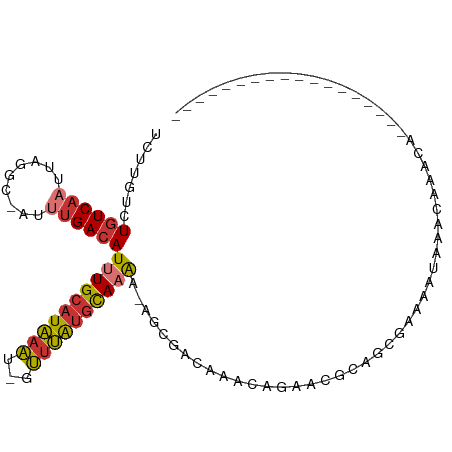
Download alignment: ClustalW | MAF
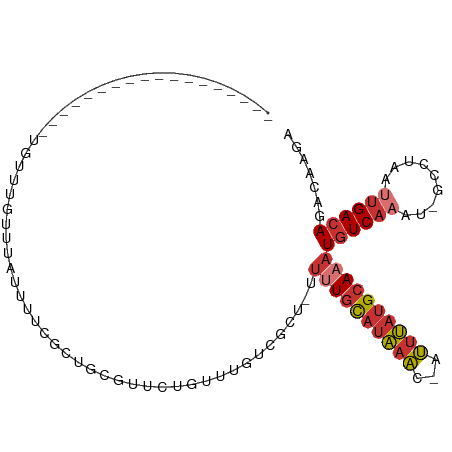
>dm3.chr3R 22732399 85 - 27905053 UCUUGUCUGUCAAUUAGGC-AUUUGACAUUUGCAUAAAU-GUUUAUGCAAAA-AGCGUCAAACAGAACGCAGCGAAAAUAAACAAACA------------------ ...((((((.....)))))-)((((...(((((((((..-..))))))))).-.((((........))))............))))..------------------ ( -18.70, z-score = -1.75, R) >droEre2.scaffold_4820 5162647 85 + 10470090 UCUUGUCUGUCAAUUAGGC-AUUUGACAUUUGCAUAAAU-GUUUAUGCAAAA-AGCGACAAACAGAACGCAGCAAAAAUAAACAAACA------------------ ..(((((((((((......-..))))))(((((((((..-..))))))))).-...)))))...........................------------------ ( -18.30, z-score = -1.97, R) >droYak2.chr3R 5076822 85 + 28832112 UCUUGUCUGUCAAUUAGGC-AUUUGACAUUUGCAUAAAU-GUUUAUGCAAAA-AGCGACAAACAGAACGCAGCGAAAAUAAACAAACA------------------ ..(((((((((((......-..))))))(((((((((..-..))))))))).-...)))))...........................------------------ ( -18.30, z-score = -1.75, R) >droSec1.super_13 1505512 85 - 2104621 UCUUGUCUGUCAAUUAGGC-AUUUGACAUUUGCAUAAAU-GUUUAUGCAAAA-AGCGACAAACAGAACGCAGCGAAAAUAAACAAACA------------------ ..(((((((((((......-..))))))(((((((((..-..))))))))).-...)))))...........................------------------ ( -18.30, z-score = -1.75, R) >droSim1.chr3R 22563249 85 - 27517382 UCUUGUCUGUCAAUUAGGC-AUUUGACAUUUGCAUAAAU-GUUUAUGCAAAA-AGCGACAAACAGAACGCAGCGAAAAUAAACAAACA------------------ ..(((((((((((......-..))))))(((((((((..-..))))))))).-...)))))...........................------------------ ( -18.30, z-score = -1.75, R) >droAna3.scaffold_13340 7100239 104 + 23697760 UCUUGUCUGUCAAUUAGGC-AUUUGACAUUUGCAUAAAU-GUUUAUGCAAAAAACAGUCAGCGACAAAACCAGAGACCAGAGCCAAAAAAACGACUAAAAUAAACA (((.(((((((((......-..))))).(((((((((..-..))))))))).....(((...)))........)))).)))......................... ( -19.60, z-score = -1.88, R) >droWil1.scaffold_181108 510141 81 - 4707319 UCUUGUUUGUCAAUUAGGCAAUGGGAAAUUUGCAUAAAU-GUUUAUGCAUAACAAAAAAAAACAGAAAAAAAAGAGCUAAAG------------------------ (((..((.(((.....)))))..)))....(((((((..-..))))))).................................------------------------ ( -12.30, z-score = -1.03, R) >droVir3.scaffold_12855 2528361 74 - 10161210 -------UGUCAAUUAAAC-AU-UGACAUUUGCGUGCCUGGGUCGGAUACGUUCUAAGCGAACAACGCACAAAUAAACAUAAA----------------------- -------(((((((.....-))-)))))..(((((((.(((..((....))..))).)).....)))))..............----------------------- ( -16.30, z-score = -1.51, R) >consensus UCUUGUCUGUCAAUUAGGC_AUUUGACAUUUGCAUAAAU_GUUUAUGCAAAA_AGCGACAAACAGAACGCAGCGAAAAUAAACAAACA__________________ .......((((((.........))))))((((((((((...))))))))))....................................................... ( -9.35 = -9.71 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:55 2011