
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,759,027 – 8,759,090 |
| Length | 63 |
| Max. P | 0.684202 |

| Location | 8,759,027 – 8,759,090 |
|---|---|
| Length | 63 |
| Sequences | 12 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 88.56 |
| Shannon entropy | 0.24292 |
| G+C content | 0.36423 |
| Mean single sequence MFE | -15.45 |
| Consensus MFE | -11.54 |
| Energy contribution | -11.69 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
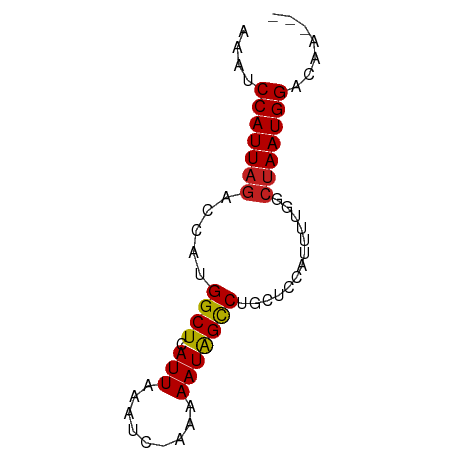
>dm3.chr2L 8759027 63 + 23011544 ---UUGUCCAUUAGCCAAAAUGGAGCAGACUAUUUUU-GAUCUAAUGAGCCAUGCUCAAAUGGUUUU ---.....((((((..(((((((......))))))).-...))))))((((((......)))))).. ( -13.40, z-score = -0.95, R) >droSim1.chr2L 8537884 63 + 22036055 ---UUGUCCAUUAGCCAAAAUGGAGCAGACUAUUUUU-GAUCUAAUGAGCCAUGCUCUAAUGGUUUU ---.....((((((..(((((((......))))))).-...))))))((((((......)))))).. ( -13.40, z-score = -0.99, R) >droSec1.super_3 4232485 63 + 7220098 ---UUGUCCAUUAGCCAAAAUGGAGCAGACUAUUUUU-GAUCUAAUGAGCCGUGCUCUAAUGGUUUU ---.....((((((..(((((((......))))))).-...))))))((((((......)))))).. ( -13.00, z-score = -0.84, R) >droYak2.chr2L 11401854 63 + 22324452 ---UUGUCCAUUAGCCAAAAUGGAGUAGACUAUUUUU-GAUUUAAUGAGCCGUGUUCUAAUGGUUUU ---...((((((......)))))).............-........(((((((......))))))). ( -11.70, z-score = -1.19, R) >droEre2.scaffold_4929 9345971 62 + 26641161 ---UUGUCCAUUAGCCAAAAUGGAGCAGACUAUUUU--GAUUUAAUGAGCCAUGGUCUAAUGGUUGU ---.....((((((.((((((((......)))))))--)..))))))((((((......)))))).. ( -15.30, z-score = -1.58, R) >droAna3.scaffold_12916 10398938 64 + 16180835 ---UUGUCCAUUAGCCAAAAUGGAGUAGGCCAUUUUUCCAUUUAAUGAGCCCUCAAAUAAUGGAUUU ---..((((((((.((.....)).(..(((((((.........)))).)))..)...)))))))).. ( -15.90, z-score = -2.12, R) >dp4.chr4_group5 1544021 63 - 2436548 ---UUGUCCAUUAGCCAAAAUGGAGCAGGCUAUUUUU-GAUUUAAUGAGCCAUGGUCUAAUGGAUUU ---..((((((((((((...(((..((....(((...-)))....))..)))))).))))))))).. ( -18.30, z-score = -2.24, R) >droPer1.super_5 1512994 63 - 6813705 ---UUGUCCAUUAGCCAAAAUGGAGCAGGCUAUUUUU-GAUUUAAUGAGCCAUGGUCUAAUGGAUUU ---..((((((((((((...(((..((....(((...-)))....))..)))))).))))))))).. ( -18.30, z-score = -2.24, R) >droWil1.scaffold_180772 48724 62 + 8906247 ---UUGUCCAUUAGU-AAAAUGGAGCAGGCUAUUUUU-GAUUUAAUGAGCCAUGGACUAAUGGAUUU ---..((((((((((-...((((..((....(((...-)))....))..))))..)))))))))).. ( -19.30, z-score = -3.76, R) >droVir3.scaffold_12723 4753220 61 + 5802038 ----UGUCCAUUAGC-AAAAUGGAGCAGGCUAUUUCU-GAUUUAAUGAGCCAGAGACUAAUGGAUUU ----.(((((((((.-....(((..((....(((...-)))....))..)))....))))))))).. ( -15.00, z-score = -1.62, R) >droMoj3.scaffold_6500 10721235 65 + 32352404 CGGCUGUCCAUUAGC-GAAAUGGAGCAGGCUAUUUUU-GAUUUAAUGAGCCAUAGUCUAAUGGAUUU .....((((((((((-...((((..((....(((...-)))....))..)))).).))))))))).. ( -15.90, z-score = -1.11, R) >droGri2.scaffold_15126 7754172 60 + 8399593 ----UGUCCAUUAGC--AAAUGGAGCAGGCUAUUUUU-GAUUUAAUGAGCCAUAGACUAAUGGAUUU ----.(((((((((.--..((((..((....(((...-)))....))..))))...))))))))).. ( -15.90, z-score = -2.47, R) >consensus ___UUGUCCAUUAGCCAAAAUGGAGCAGGCUAUUUUU_GAUUUAAUGAGCCAUGGUCUAAUGGAUUU .......(((((((.....((((..((....(((....)))....))..))))...))))))).... (-11.54 = -11.69 + 0.15)



| Location | 8,759,027 – 8,759,090 |
|---|---|
| Length | 63 |
| Sequences | 12 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 88.56 |
| Shannon entropy | 0.24292 |
| G+C content | 0.36423 |
| Mean single sequence MFE | -14.19 |
| Consensus MFE | -8.16 |
| Energy contribution | -8.01 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.592227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8759027 63 - 23011544 AAAACCAUUUGAGCAUGGCUCAUUAGAUC-AAAAAUAGUCUGCUCCAUUUUGGCUAAUGGACAA--- ....(((((.(((((.((((.(((.....-...))))))))))))..........)))))....--- ( -12.20, z-score = -0.67, R) >droSim1.chr2L 8537884 63 - 22036055 AAAACCAUUAGAGCAUGGCUCAUUAGAUC-AAAAAUAGUCUGCUCCAUUUUGGCUAAUGGACAA--- ....(((((((((((.((((.(((.....-...)))))))))))((.....)))))))))....--- ( -15.50, z-score = -2.12, R) >droSec1.super_3 4232485 63 - 7220098 AAAACCAUUAGAGCACGGCUCAUUAGAUC-AAAAAUAGUCUGCUCCAUUUUGGCUAAUGGACAA--- ....(((((((((((.((((.(((.....-...)))))))))))((.....)))))))))....--- ( -15.50, z-score = -2.51, R) >droYak2.chr2L 11401854 63 - 22324452 AAAACCAUUAGAACACGGCUCAUUAAAUC-AAAAAUAGUCUACUCCAUUUUGGCUAAUGGACAA--- ....(((((((.....((((.(((.....-...)))))))....((.....)))))))))....--- ( -11.10, z-score = -2.59, R) >droEre2.scaffold_4929 9345971 62 - 26641161 ACAACCAUUAGACCAUGGCUCAUUAAAUC--AAAAUAGUCUGCUCCAUUUUGGCUAAUGGACAA--- ....(((((((.(((.(((..((((....--....))))..)))......))))))))))....--- ( -14.30, z-score = -2.46, R) >droAna3.scaffold_12916 10398938 64 - 16180835 AAAUCCAUUAUUUGAGGGCUCAUUAAAUGGAAAAAUGGCCUACUCCAUUUUGGCUAAUGGACAA--- ...(((((((.....((((.((((.........))))))))...((.....)).)))))))...--- ( -14.00, z-score = -0.94, R) >dp4.chr4_group5 1544021 63 + 2436548 AAAUCCAUUAGACCAUGGCUCAUUAAAUC-AAAAAUAGCCUGCUCCAUUUUGGCUAAUGGACAA--- ...((((((((.(((.((((.(((.....-...)))))))..........)))))))))))...--- ( -16.80, z-score = -3.11, R) >droPer1.super_5 1512994 63 + 6813705 AAAUCCAUUAGACCAUGGCUCAUUAAAUC-AAAAAUAGCCUGCUCCAUUUUGGCUAAUGGACAA--- ...((((((((.(((.((((.(((.....-...)))))))..........)))))))))))...--- ( -16.80, z-score = -3.11, R) >droWil1.scaffold_180772 48724 62 - 8906247 AAAUCCAUUAGUCCAUGGCUCAUUAAAUC-AAAAAUAGCCUGCUCCAUUUU-ACUAAUGGACAA--- ...(((((((((.((.((((.(((.....-...))))))))).........-)))))))))...--- ( -13.10, z-score = -3.15, R) >droVir3.scaffold_12723 4753220 61 - 5802038 AAAUCCAUUAGUCUCUGGCUCAUUAAAUC-AGAAAUAGCCUGCUCCAUUUU-GCUAAUGGACA---- ...(((((((((....((((.(((.....-...)))))))...........-)))))))))..---- ( -13.26, z-score = -1.90, R) >droMoj3.scaffold_6500 10721235 65 - 32352404 AAAUCCAUUAGACUAUGGCUCAUUAAAUC-AAAAAUAGCCUGCUCCAUUUC-GCUAAUGGACAGCCG ...((((((((...((((..((.......-..........))..))))...-.))))))))...... ( -12.63, z-score = -1.96, R) >droGri2.scaffold_15126 7754172 60 - 8399593 AAAUCCAUUAGUCUAUGGCUCAUUAAAUC-AAAAAUAGCCUGCUCCAUUU--GCUAAUGGACA---- ...(((((((((..((((..((.......-..........))..))))..--)))))))))..---- ( -15.13, z-score = -3.31, R) >consensus AAAUCCAUUAGACCAUGGCUCAUUAAAUC_AAAAAUAGCCUGCUCCAUUUUGGCUAAUGGACAA___ ....(((((((.....((((.(((.........))))))).............)))))))....... ( -8.16 = -8.01 + -0.15)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:53 2011