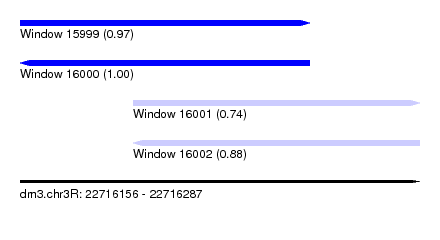
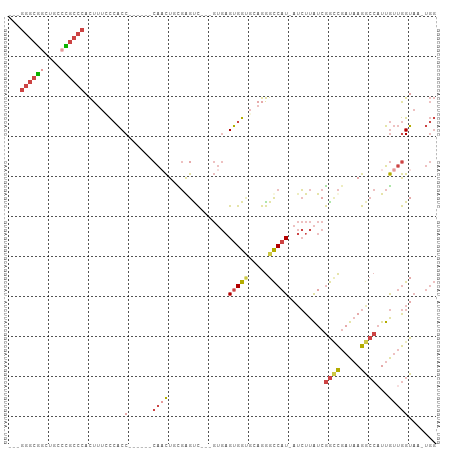
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,716,156 – 22,716,287 |
| Length | 131 |
| Max. P | 0.998551 |

| Location | 22,716,156 – 22,716,251 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 69.86 |
| Shannon entropy | 0.57639 |
| G+C content | 0.60679 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -17.68 |
| Energy contribution | -19.74 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
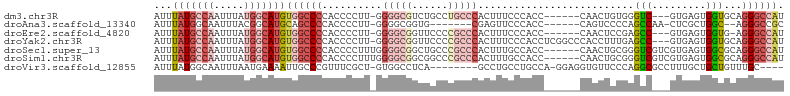
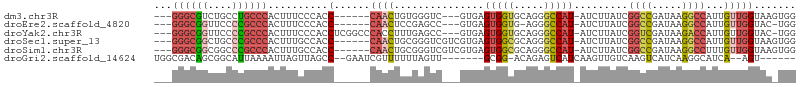
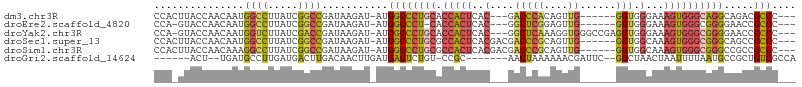
>dm3.chr3R 22716156 95 + 27905053 AUUUAUGCCAAUUUAUGGCAUGUGGCCCCACCCCUU-GGGGCGUCUGCCUGCCCACUUUCCCACC------CAACUGUGGGUC---GUGAGUGGUGCAGGGCCAU ...(((((((.....)))))))..((((((.....)-))))).....(((((((((((.(((((.------.....)))))..---..)))))).)))))..... ( -46.30, z-score = -3.32, R) >droAna3.scaffold_13340 7084330 88 - 23697760 AUUUAUGGCAAUUUACGGCAUGCAGCCCCACCCCUU-GGGGCGGUG-------CGAGUUCCCACC------CAGUCCCCAGCCAA-CUCGCUGGC--AGGGCCGC ...............((((.(((.((((((.....)-)))))((((-------.(....).))))------.......((((...-...))))))--)..)))). ( -32.30, z-score = -0.08, R) >droEre2.scaffold_4820 5145909 94 - 10470090 AUUUAUGCCAAUUUAUGGCAUGUGGCCCCACCCCUU-GGGGCGGUUCCCCGCCCACUUUCCCACC------CAACUCCGAGCC---GUGAGUGGUG-AGGGCCAU ...(((((((.....)))))))((((((((((....-.((((((....))))))...........------..((((((...)---).))))))))-.)))))). ( -43.20, z-score = -3.71, R) >droYak2.chr3R 5059706 101 - 28832112 AUUUAUGCCAAUUUAUGGCAUGUGGCCCCACCCCUU-GGGGCGGUUCCCCGCCCACUUUCCCACCUCGGCCCACCUUUGAGCC---GUGAGUGGUGCAGGGCCAU ...(((((((.....)))))))((((((........-.((((((....))))))......(((((.((((.((....)).)))---).).))))....)))))). ( -46.70, z-score = -3.41, R) >droSec1.super_13 1489458 99 + 2104621 AUUUAUGCCAAUUUAUGGCAUGUGGCCCCACCCCUUUGGGGCGGCUGCCCGCCCACUUUGCCACC------CAACUGCGGGUCGUCGUGAGUGGCGCAGGGCCAU ....((((((.....))))))((.((((((......)))))).)).((((((((((((.((.(((------(......)))).))...)))))).)).))))... ( -48.20, z-score = -2.34, R) >droSim1.chr3R 22547059 99 + 27517382 AUUUAUGCCAAUUUAUGGCAUGUGGCCCCACCCCUUUGGGGCGGCGGCCCGCCCACUUUGCCACC------CAACUGCGGGUCGUCGUGAGUGGCGCAGGGCCAU ....((((((.....))))))((.((((((......)))))).))(((((((((((((.((.(((------(......)))).))...)))))).)).))))).. ( -53.20, z-score = -3.28, R) >droVir3.scaffold_12855 2501726 91 + 10161210 AUUUAUGGCAAUUUAAUGAAAAUUGCCCGUUUCGCU-GUGGCCUCA--------GCCUGCCUGCCA-GGAGGUGUUCCCAGGCGCCUUUGCUGCUGUUUGC---- ......((((((((.....))))))))......(((-(......))--------))..((..((..-((((((((......)))))))))).)).......---- ( -25.30, z-score = 0.55, R) >consensus AUUUAUGCCAAUUUAUGGCAUGUGGCCCCACCCCUU_GGGGCGGCUGCCCGCCCACUUUCCCACC______CAACUCCGAGCC___GUGAGUGGUGCAGGGCCAU ...(((((((.....)))))))((((((..........(((((......)))))..........................(((.........)))...)))))). (-17.68 = -19.74 + 2.06)
| Location | 22,716,156 – 22,716,251 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 69.86 |
| Shannon entropy | 0.57639 |
| G+C content | 0.60679 |
| Mean single sequence MFE | -47.46 |
| Consensus MFE | -22.03 |
| Energy contribution | -24.69 |
| Covariance contribution | 2.66 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.998551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
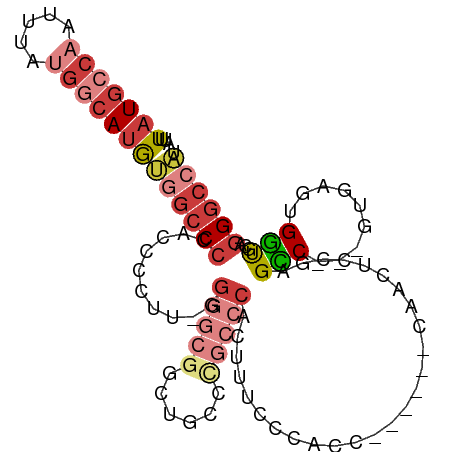
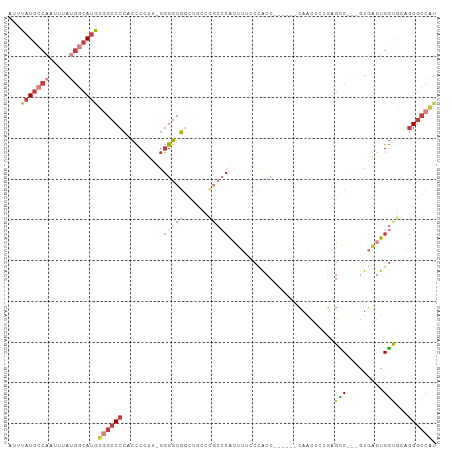
>dm3.chr3R 22716156 95 - 27905053 AUGGCCCUGCACCACUCAC---GACCCACAGUUG------GGUGGGAAAGUGGGCAGGCAGACGCCCC-AAGGGGUGGGGCCACAUGCCAUAAAUUGGCAUAAAU .(((((((((.(((((..(---.(((((....))------))).)...))))))))......(((((.-...))))))))))).((((((.....)))))).... ( -49.60, z-score = -3.86, R) >droAna3.scaffold_13340 7084330 88 + 23697760 GCGGCCCU--GCCAGCGAG-UUGGCUGGGGACUG------GGUGGGAACUCG-------CACCGCCCC-AAGGGGUGGGGCUGCAUGCCGUAAAUUGCCAUAAAU (((((((.--.(((((...-...)))((((....------((((.(....).-------)))).))))-....))..)))))))(((.((.....)).))).... ( -41.00, z-score = -0.97, R) >droEre2.scaffold_4820 5145909 94 + 10470090 AUGGCCCU-CACCACUCAC---GGCUCGGAGUUG------GGUGGGAAAGUGGGCGGGGAACCGCCCC-AAGGGGUGGGGCCACAUGCCAUAAAUUGGCAUAAAU .((((((.-((((.(((((---(((.....))).------.)))))...(.((((((....)))))))-....)))))))))).((((((.....)))))).... ( -48.60, z-score = -3.55, R) >droYak2.chr3R 5059706 101 + 28832112 AUGGCCCUGCACCACUCAC---GGCUCAAAGGUGGGCCGAGGUGGGAAAGUGGGCGGGGAACCGCCCC-AAGGGGUGGGGCCACAUGCCAUAAAUUGGCAUAAAU .((((((..(.(((((..(---((((((....))))))).)))))....(.((((((....)))))))-.....)..)))))).((((((.....)))))).... ( -58.70, z-score = -5.13, R) >droSec1.super_13 1489458 99 - 2104621 AUGGCCCUGCGCCACUCACGACGACCCGCAGUUG------GGUGGCAAAGUGGGCGGGCAGCCGCCCCAAAGGGGUGGGGCCACAUGCCAUAAAUUGGCAUAAAU .(((((((((.((.(((((..(.(((((....))------))).)....))))).))))))(((((((...)))))))))))).((((((.....)))))).... ( -52.80, z-score = -3.16, R) >droSim1.chr3R 22547059 99 - 27517382 AUGGCCCUGCGCCACUCACGACGACCCGCAGUUG------GGUGGCAAAGUGGGCGGGCCGCCGCCCCAAAGGGGUGGGGCCACAUGCCAUAAAUUGGCAUAAAU .((((((..((((((((((..((...))..).))------))))))...(.((((((....)))))))......)..)))))).((((((.....)))))).... ( -52.30, z-score = -2.66, R) >droVir3.scaffold_12855 2501726 91 - 10161210 ----GCAAACAGCAGCAAAGGCGCCUGGGAACACCUCC-UGGCAGGCAGGC--------UGAGGCCAC-AGCGAAACGGGCAAUUUUCAUUAAAUUGCCAUAAAU ----((.....((.((....))(((.((((.....)))-))))..)).(((--------....)))..-.))......((((((((.....))))))))...... ( -29.20, z-score = -1.19, R) >consensus AUGGCCCUGCACCACUCAC___GACUCGCAGUUG______GGUGGGAAAGUGGGCGGGCAGCCGCCCC_AAGGGGUGGGGCCACAUGCCAUAAAUUGGCAUAAAU .((((((....(((((......(((.....))).......))))).................((((((...)))))))))))).((((((.....)))))).... (-22.03 = -24.69 + 2.66)

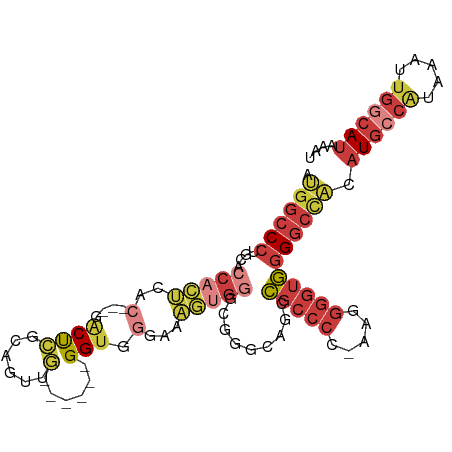
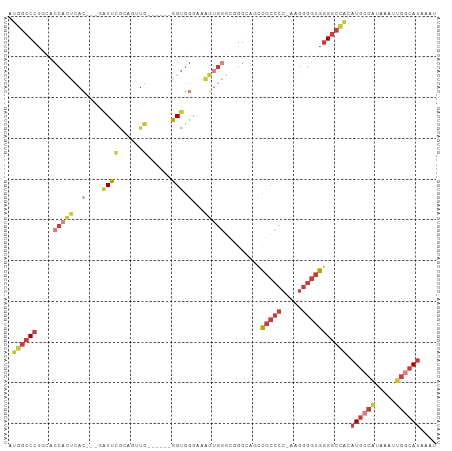
| Location | 22,716,193 – 22,716,287 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 69.13 |
| Shannon entropy | 0.55244 |
| G+C content | 0.58079 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -17.77 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
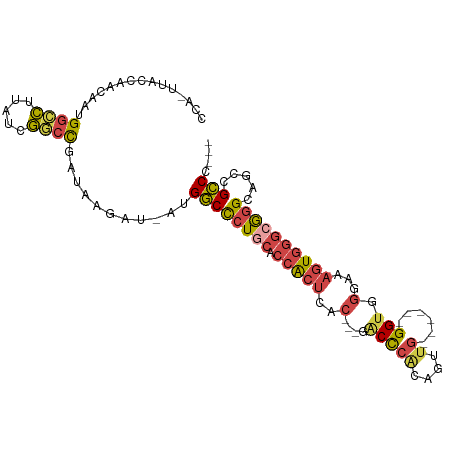
>dm3.chr3R 22716193 94 + 27905053 ---GGGCGUCUGCCUGCCCACUUUCCCACC------CAACUGUGGGUC---GUGAGUGGUGCAGGGCCAU-AUCUUAUCGGCCGAUAAGGCCAUUGUUGGUAAGUGG ---.(((.....(((((((((((.(((((.------.....)))))..---..)))))).))))))))..-..((((((((((.....))))......))))))... ( -39.60, z-score = -1.93, R) >droEre2.scaffold_4820 5145946 92 - 10470090 ---GGGCGGUUCCCCGCCCACUUUCCCACC------CAACUCCGAGCC---GUGAGUGGUG-AGGGCCAU-AUCUUAUCGGCCGAUAAGGCCAUUGUUGGUAC-UGG ---((((((....))))))....(((((((------..((((((...)---).))))))))-.)))((((-(((.((..((((.....))))..))..)))).-))) ( -35.90, z-score = -1.40, R) >droYak2.chr3R 5059743 99 - 28832112 ---GGGCGGUUCCCCGCCCACUUUCCCACCUCGGCCCACCUUUGAGCC---GUGAGUGGUGCAGGGCCAU-AUCUUAUCGGUCGAUAAGACCAUUGUUGGUAC-UGG ---((((((....))))))......(((((.((((.((....)).)))---).).))))..(((..(((.-.(((((((....))))))).......)))..)-)). ( -40.60, z-score = -2.46, R) >droSec1.super_13 1489496 97 + 2104621 ---GGGCGGCUGCCCGCCCACUUUGCCACC------CAACUGCGGGUCGUCGUGAGUGGCGCAGGGCCAU-AUCUUAUCGGCCGAUAAGGCCAUUGUUGGUAAGUGG ---((((((....))))))......(((((------((((...(.((((.(....))))).)..((((.(-(((.........)))).))))...)))))...)))) ( -40.00, z-score = -0.50, R) >droSim1.chr3R 22547097 97 + 27517382 ---GGGCGGCGGCCCGCCCACUUUGCCACC------CAACUGCGGGUCGUCGUGAGUGGCGCAGGGCCAU-AUCUUAUCGGCCGAUAAGGCCUUUGUUGGUAAGUGG ---.(.(((((((((((((((((.((.(((------(......)))).))...)))))).)).)))))..-........((((.....))))...)))).)...... ( -41.90, z-score = -0.79, R) >droGri2.scaffold_14624 1799435 89 - 4233967 UGGCGACAGCGGCAUUAAAAUUAGUUAGCC--GAAUCGUUUUUUAGUU-------GCGG-ACAGAGUCAUCAAGUUGUCAAGUCAUCAAGGCAUCA--AGU------ (((((((..((((((((....))))..)))--)..((((.........-------))))-.............))))))).(((.....)))....--...------ ( -16.90, z-score = -0.55, R) >consensus ___GGGCGGCUGCCCGCCCACUUUCCCACC______CAACUGCGAGUC___GUGAGUGGUGCAGGGCCAU_AUCUUAUCGGCCGAUAAGGCCAUUGUUGGUAA_UGG ...((((((....))))))........(((.........................(((((.....))))).........((((.....))))......)))...... (-17.77 = -18.47 + 0.70)
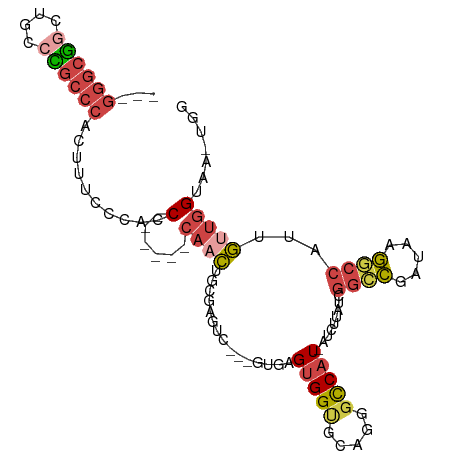
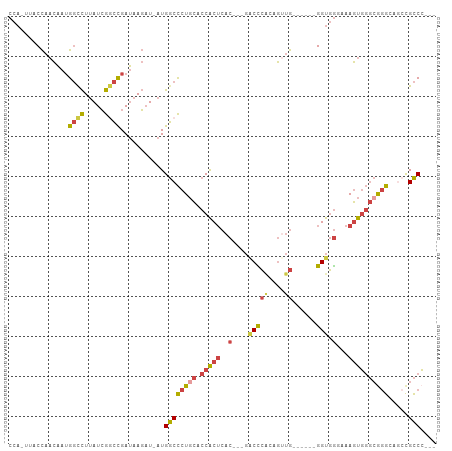
| Location | 22,716,193 – 22,716,287 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.13 |
| Shannon entropy | 0.55244 |
| G+C content | 0.58079 |
| Mean single sequence MFE | -34.49 |
| Consensus MFE | -16.41 |
| Energy contribution | -16.83 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22716193 94 - 27905053 CCACUUACCAACAAUGGCCUUAUCGGCCGAUAAGAU-AUGGCCCUGCACCACUCAC---GACCCACAGUUG------GGUGGGAAAGUGGGCAGGCAGACGCCC--- ..............(((((.....))))).......-..((((((((.(((((..(---.(((((....))------))).)...)))))))))).....))).--- ( -36.00, z-score = -2.20, R) >droEre2.scaffold_4820 5145946 92 + 10470090 CCA-GUACCAACAAUGGCCUUAUCGGCCGAUAAGAU-AUGGCCCU-CACCACUCAC---GGCUCGGAGUUG------GGUGGGAAAGUGGGCGGGGAACCGCCC--- ...-...........((((.((((.........)))-).))))((-((((((((.(---(...))))))..------)))))).....((((((....))))))--- ( -36.50, z-score = -1.11, R) >droYak2.chr3R 5059743 99 + 28832112 CCA-GUACCAACAAUGGUCUUAUCGACCGAUAAGAU-AUGGCCCUGCACCACUCAC---GGCUCAAAGGUGGGCCGAGGUGGGAAAGUGGGCGGGGAACCGCCC--- .((-(..(((......((((((((....))))))))-.)))..)))..(((((..(---((((((....))))))).)))))......((((((....))))))--- ( -45.40, z-score = -3.33, R) >droSec1.super_13 1489496 97 - 2104621 CCACUUACCAACAAUGGCCUUAUCGGCCGAUAAGAU-AUGGCCCUGCGCCACUCACGACGACCCGCAGUUG------GGUGGCAAAGUGGGCGGGCAGCCGCCC--- ((((((.........((((.((((.........)))-).))))....(((((((((..((...))..).))------)))))).))))))..((((....))))--- ( -38.50, z-score = -1.30, R) >droSim1.chr3R 22547097 97 - 27517382 CCACUUACCAACAAAGGCCUUAUCGGCCGAUAAGAU-AUGGCCCUGCGCCACUCACGACGACCCGCAGUUG------GGUGGCAAAGUGGGCGGGCCGCCGCCC--- ...............(((((((((....))))))..-..(((((.((.(((((...(.(.(((((....))------))).))..)))))))))))))))....--- ( -39.30, z-score = -1.44, R) >droGri2.scaffold_14624 1799435 89 + 4233967 ------ACU--UGAUGCCUUGAUGACUUGACAACUUGAUGACUCUGU-CCGC-------AACUAAAAAACGAUUC--GGCUAACUAAUUUUAAUGCCGCUGUCGCCA ------...--.......(((..(((..(.((......)).)...))-)..)-------))........((((.(--(((..............))))..))))... ( -11.24, z-score = -0.19, R) >consensus CCA_UUACCAACAAUGGCCUUAUCGGCCGAUAAGAU_AUGGCCCUGCACCACUCAC___GACCCACAGUUG______GGUGGGAAAGUGGGCGGGCAGCCGCCC___ .....((((......((((.....))))..........(((.......)))..........................)))).......((((((....))))))... (-16.41 = -16.83 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:52 2011