
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,673,780 – 22,673,838 |
| Length | 58 |
| Max. P | 0.693824 |

| Location | 22,673,780 – 22,673,838 |
|---|---|
| Length | 58 |
| Sequences | 10 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 76.80 |
| Shannon entropy | 0.51369 |
| G+C content | 0.36692 |
| Mean single sequence MFE | -10.82 |
| Consensus MFE | -4.81 |
| Energy contribution | -5.60 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22673780 58 - 27905053 ---AAAAACUGUCUGCGAGAUAAUGCGGUAAAAUAAUCUUGCUGCACUUGGGAAAAUCUCU ---.............(((((..((((((((.......))))))))(....)...))))). ( -11.50, z-score = -2.06, R) >droSim1.chr3R 22504239 58 - 27517382 ---AAAAACUGUCUGCGAGAUAAUGCGGUAAAAUAAUCUUGCUGCACUUGGGAAAAUCUCU ---.............(((((..((((((((.......))))))))(....)...))))). ( -11.50, z-score = -2.06, R) >droSec1.super_13 1447256 58 - 2104621 ---AAAAACUGUCUGCGAGAUAAUGCGGUAAAAUAAUCUUGCUGCACUUGGGAAAAUCUCU ---.............(((((..((((((((.......))))))))(....)...))))). ( -11.50, z-score = -2.06, R) >droEre2.scaffold_4820 5102166 58 + 10470090 ---AAAAACUGUCUGCGAGAUAAUGCGGUAAAAUAAUCUUGCUGCACUUGGGAAAAUCUCU ---.............(((((..((((((((.......))))))))(....)...))))). ( -11.50, z-score = -2.06, R) >droAna3.scaffold_13340 7038822 54 + 23697760 ----AAAACUGU---CGAGAUAAUGCGGUAAAAUAAUCUAGCUGCUCUUGGGAAAAUCUCU ----........---.(((((...(((((...........)))))((....))..))))). ( -8.90, z-score = -1.11, R) >dp4.chr2 25996984 58 + 30794189 ---AAAAACUGUCUACGAGAUAAUGCGGUAAAAUAAUCUUGCUGCACUUGAGAAUAUCUCU ---.............((((((.((((((((.......))))))))(....)..)))))). ( -12.80, z-score = -3.20, R) >droPer1.super_19 1375595 58 + 1869541 ---AAAAACUGUCUACGAGAUAAUGCGGUAAAAUAAUCUUGCUGCACUUGAGAAUAUCUCU ---.............((((((.((((((((.......))))))))(....)..)))))). ( -12.80, z-score = -3.20, R) >droWil1.scaffold_181130 14011248 50 + 16660200 ---AAAACUUGUCUACAAGAUAAUGCGGUAAAAUAAUCUAGCUGAACUUGAGA-------- ---........(((.((((......((((...........))))..)))))))-------- ( -4.60, z-score = 0.19, R) >droMoj3.scaffold_6540 4372531 58 + 34148556 UAAUGUCAUUGUCG---UGAUAAUGCGGUUAAAUAAUCUCCUUGGAAACAAGAGCGUUUGA ...((((((....)---)))))((((((((....))))..((((....)))).)))).... ( -13.30, z-score = -1.93, R) >droGri2.scaffold_14624 2170950 61 - 4233967 AAAUAUAAUUGUCGAGCAGAUAAAGACGUCAAAUAAUCGGGCUGCAAAUGCGAGCGCUUCU .............((((.(((......)))..........(((((....)).))))))).. ( -9.80, z-score = 0.67, R) >consensus ___AAAAACUGUCUACGAGAUAAUGCGGUAAAAUAAUCUUGCUGCACUUGGGAAAAUCUCU .........((((.....))))..(((((((.......)))))))................ ( -4.81 = -5.60 + 0.79)
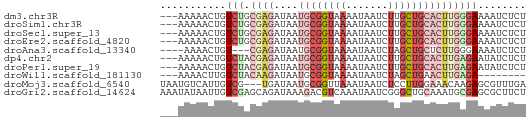

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:45 2011