| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,634,527 – 22,634,623 |
| Length | 96 |
| Max. P | 0.978827 |

| Location | 22,634,527 – 22,634,623 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 68.54 |
| Shannon entropy | 0.64621 |
| G+C content | 0.40248 |
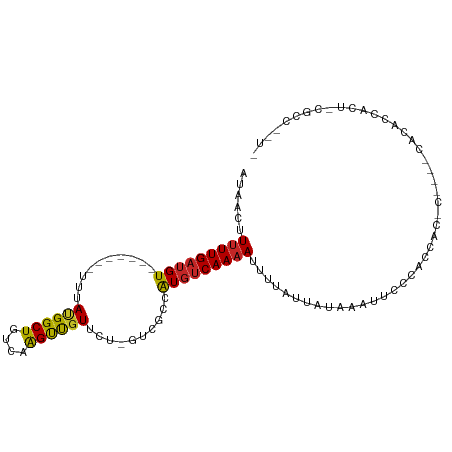
| Mean single sequence MFE | -14.74 |
| Consensus MFE | -9.29 |
| Energy contribution | -9.03 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.978827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
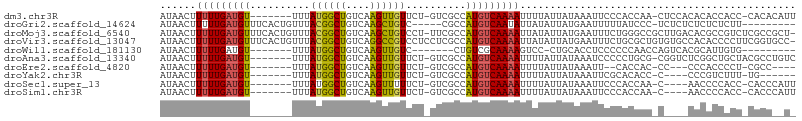
>dm3.chr3R 22634527 96 - 27905053 AUAACUUUUUGAUGU-------UUUAUGGCUGUCAAGUUGUUCU-GUCGCCAUGUCAAAAUUUUAUUAUAAAUUCCCACCAA-CUCCACACACCACC-CACACAUU ((((..(((((((..-------...(((((.(.((........)-).)))))))))))))..))))................-..............-........ ( -11.30, z-score = -0.34, R) >droGri2.scaffold_14624 2106755 91 - 4233967 AUAACUUUUUGAUGUUUCACUGUUUACGGCUGUCAAGCUGUC-----CGCCAUGUCAAUAUUAUAUUAUGAAUUUUUAUCCC-UCUCUCUCUCUCUU--------- .........(((((((.((..((..((((((....)))))).-----.))..))..)))))))...................-..............--------- ( -11.80, z-score = -1.92, R) >droMoj3.scaffold_6540 4326226 104 + 34148556 AUAACUUUUUGAUGUUUCACUGUUUACGGCUGUCAAGCUGUCCU-UUCGCCAUGUCAAAAUUAUAUUAUGAAUUUCUGGGCCGCUUGACACGCCGUCUCGCCGCU- .........(((....)))......(((((((((((((.((((.-((((..((((.......))))..)))).....)))).)))))))).))))).........- ( -28.20, z-score = -2.90, R) >droVir3.scaffold_13047 11952327 105 - 19223366 AUAACUUUUUGAUGUUUCACUGUUUACGGCUGUCAGGCCGUCCUCCUCGCCAUGUCAAAAUUAUAUUAUGAAUUUCUGCGCUGUGUGCCACACCCCUUCGGUGCC- ......(((((((((..........((((((....))))))..........))))))))).................((((((.(((...))).....)))))).- ( -20.85, z-score = -0.54, R) >droWil1.scaffold_181130 13954666 82 + 16660200 AUAACUUUUUGAUGU-------UUUAUGGCUGUCAAGUUGUC-------CUGUCGCAAAAGUCC-CUGCACCUCCCCCCAACCAGUCACGCAUUGUG--------- ..........(((((-------....((((((....((((..-------..(..(((.......-.)))..)......)))))))))).)))))...--------- ( -11.70, z-score = -0.07, R) >droAna3.scaffold_13340 6999392 97 + 23697760 AUAACUUUUUGAUGU-------UUUAUGGCUGUCAAGUUGUUCU-GUCGCCAUGUCAAAAUUUUAUUAUAAAUCCCCCUGCG-CGGUCUCGGCUGCUACGCCUGUC ((((..(((((((..-------...(((((.(.((........)-).)))))))))))))..))))...............(-((((....))))).......... ( -18.30, z-score = -0.74, R) >droEre2.scaffold_4820 5061743 87 + 10470090 AUAACUUUUUGAUGU-------UUUAUGGCUGUCAAGUUGUUCU-GUCGCCAUGUCAAAAUUUUAUUAUAAAUU--CACCAC-CC---CCCACCCCU-CGCC---- ((((..(((((((..-------...(((((.(.((........)-).)))))))))))))..))))........--......-..---.........-....---- ( -11.30, z-score = -1.25, R) >droYak2.chr3R 4974065 86 + 28832112 AUAACUUUUUGAUGU-------UUUAUGGCUGUCAAGUUGUUCU-GUCGCCAUGUCAAAAUUUUAUUAUAAAUUCGCACACC-C----CCCGUCUUU-UG------ ((((..(((((((..-------...(((((.(.((........)-).)))))))))))))..))))................-.----.........-..------ ( -11.30, z-score = -0.94, R) >droSec1.super_13 1408820 92 - 2104621 AUAACUUUUUGAUGU-------UUUAUGGCUGUCAAGUUUUUCU-GUCGCCAUGUCAAAAUUUUAUUAUAAAUUCCCACCAA-C----AACCCCACC-CACCCAUU ((((..(((((((..-------...(((((.(.((........)-).)))))))))))))..))))................-.----.........-........ ( -11.30, z-score = -1.49, R) >droSim1.chr3R 22466165 92 - 27517382 AUAACUUUUUGAUGU-------UUUAUGGCUGUCAAGUUGUUCU-GUCGCCAUGUCAAAAUUUUAUUAUAAAUUCCCACCAA-C----AACCCCACC-CACCCAUU ((((..(((((((..-------...(((((.(.((........)-).)))))))))))))..))))................-.----.........-........ ( -11.30, z-score = -0.91, R) >consensus AUAACUUUUUGAUGU_______UUUAUGGCUGUCAAGUUGUUCU_GUCGCCAUGUCAAAAUUUUAUUAUAAAUUCCCACCAC_C____CACACCACU_CGCC__U_ ......(((((((((..........((((((....))))))..........))))))))).............................................. ( -9.29 = -9.03 + -0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:41 2011