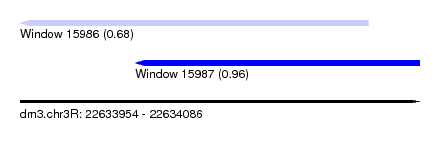
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,633,954 – 22,634,086 |
| Length | 132 |
| Max. P | 0.957870 |

| Location | 22,633,954 – 22,634,069 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.48 |
| Shannon entropy | 0.50794 |
| G+C content | 0.37543 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -10.77 |
| Energy contribution | -13.66 |
| Covariance contribution | 2.89 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
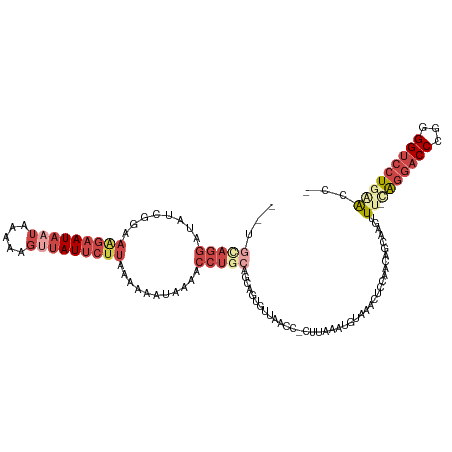
>dm3.chr3R 22633954 115 - 27905053 --UGUAGGAUAUCGGAAAGAAUAAUAAAAAGUUAUUCUUAAAAAAUAAGACCUGCAGCAGUGUUAACC-CUUAAAUGUAAACUCCAACAGCAAGUUU-CAGGACCCGGGGUCCU-CGCAC --((((((((..(((.(((((((((.....)))))))))...........((((.(((.((....)).-......(((........)))....))).-))))..)))..)))))-.))). ( -27.00, z-score = -1.99, R) >droSim1.chr3R 22465641 116 - 27517382 --UGCAGGAUACCGAAAAGAAUAAUAAAAAGCUAUUCUUAAAAAAUGAAACCUGCAGCAGUGUUAACC-CUUAAAUGUAAACUCCAACAGCAAGUUU-CAGGACCCGGGGUCCUGAACGC --..(((((..(((..(((((((.........)))))))......((((((.(((......(((.((.-.......)).))).......))).))))-)).....)))..)))))..... ( -27.22, z-score = -2.14, R) >droSec1.super_13 1408283 116 - 2104621 --UGCAGGAUAUCGAAAAGAAUAAUAAAAAGUUAUUCUUAAAAAAUGAAACCUGCAGCAGUGUUAACC-CUUAAAUGUAAACUCCAACAGCAAGUUU-GAGGACCCGGGGUCCUGAAUGC --((((((...(((..(((((((((.....)))))))))......)))..))))))(((......(((-((.....((...(((.(((.....))).-))).))..)))))......))) ( -26.90, z-score = -2.20, R) >droYak2.chr3R 4973435 115 + 28832112 --CGUAGGAAAUCGGAAAGAAUAAUAAAAAGUUAUACUUAAAAAAUAAAACCUGCAGCUGUGUUACCC-CUUAAACGUAAACUCCAACAGCAAGUUU-CAGGACCCGGGGUCCUGGAAC- --.(((((...((.....))(((((.....)))))...............))))).(((((.((((..-.......))))......)))))..((((-(((((((...)))))))))))- ( -30.10, z-score = -2.71, R) >droEre2.scaffold_4820 5061148 118 + 10470090 CUUAGAGGAAAUCGGAAGGAAUAAUAAAAAGUUAUUCUUAAAAAAUAAAACCUGCAGCUGUGUUACCCUUUUAAACACAAACUCCAACAGCAAGUUU-UGGGACCCGAGGUCCUGCAAC- .....((((..((((.(((((((((.....)))))))))......((((((.(((((.((((((.........))))))..))......))).))))-))....))))..)))).....- ( -27.70, z-score = -1.87, R) >droAna3.scaffold_13340 6998924 88 + 23697760 -------------------AAUAUUUUAAA---AUUCUUGAAAAGUUUAUAUUUUAAAAUGGA------UUUGAGGACAAACUGUUACAGAAAGCUUGCAGGACCUGAGG--AUGAAC-- -------------------..(((((((((---((...(((.....))).)))))))))))..------(((.(((.....((((...((....)).))))..))).)))--......-- ( -10.80, z-score = 0.44, R) >consensus __UGCAGGAUAUCGGAAAGAAUAAUAAAAAGUUAUUCUUAAAAAAUAAAACCUGCAGCAGUGUUAACC_CUUAAAUGUAAACUCCAACAGCAAGUUU_CAGGACCCGGGGUCCUGAACC_ ...(((((........(((((((((.....)))))))))...........)))))...........................................(((((((...)))))))..... (-10.77 = -13.66 + 2.89)

| Location | 22,633,992 – 22,634,086 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Shannon entropy | 0.31682 |
| G+C content | 0.34491 |
| Mean single sequence MFE | -18.43 |
| Consensus MFE | -12.48 |
| Energy contribution | -12.74 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.957870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
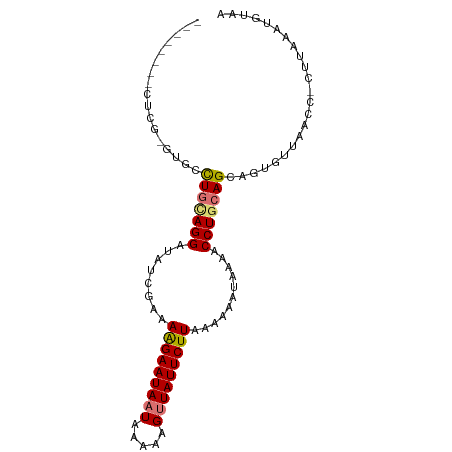
>dm3.chr3R 22633992 94 - 27905053 GUGCACACUUCG-GUGCCUGUAGGAUAUCGGAAAGAAUAAUAAAAAGUUAUUCUUAAAAAAUAAGACCUGCAGCAGUGUUAACC-CUUAAAUGUAA ..((((((....-))((.((((((.(((....(((((((((.....))))))))).....)))...)))))))).)))).....-........... ( -20.80, z-score = -2.14, R) >droSim1.chr3R 22465680 86 - 27517382 --------CUCG-UUGCCUGCAGGAUACCGAAAAGAAUAAUAAAAAGCUAUUCUUAAAAAAUGAAACCUGCAGCAGUGUUAACC-CUUAAAUGUAA --------..((-((((.((((((........(((((((.........)))))))...........))))))))))))......-........... ( -15.61, z-score = -2.21, R) >droSec1.super_13 1408322 86 - 2104621 --------CUCG-UUGCCUGCAGGAUAUCGAAAAGAAUAAUAAAAAGUUAUUCUUAAAAAAUGAAACCUGCAGCAGUGUUAACC-CUUAAAUGUAA --------..((-((((.((((((...(((..(((((((((.....)))))))))......)))..))))))))))))......-........... ( -19.20, z-score = -3.77, R) >droEre2.scaffold_4820 5061186 96 + 10470090 GUACACGCACAGUGCCCUUAGAGGAAAUCGGAAGGAAUAAUAAAAAGUUAUUCUUAAAAAAUAAAACCUGCAGCUGUGUUACCCUUUUAAACACAA (((...(((((((((((.....))........(((((((((.....)))))))))..............))).))))))))).............. ( -18.10, z-score = -1.36, R) >consensus ________CUCG_GUGCCUGCAGGAUAUCGAAAAGAAUAAUAAAAAGUUAUUCUUAAAAAAUAAAACCUGCAGCAGUGUUAACC_CUUAAAUGUAA .................(((((((........(((((((((.....)))))))))...........)))))))....................... (-12.48 = -12.74 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:40 2011