| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,626,458 – 22,626,552 |
| Length | 94 |
| Max. P | 0.948358 |

| Location | 22,626,458 – 22,626,552 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 61.71 |
| Shannon entropy | 0.75289 |
| G+C content | 0.40472 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -8.61 |
| Energy contribution | -9.32 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22626458 94 + 27905053 ----GAUUUAGGUUAAGAUUUUUCUUCUUUGGCCCACAUCAAAGGCAA--AGAAGCUUUCAAAG-CGGCUAUA-ACAAUGGCGGCAAACUGAAAAAAAAAAA- ----..(((((.....((.....((((((((.((.........)))))--)))))...))...(-(.(((((.-...))))).))...))))).........- ( -22.20, z-score = -1.65, R) >droSim1.chr3R 22458240 95 + 27517382 ----GAUUUGGGUUAAGAUUUUUCCUCUUUGGCCCAGAUCAAAGGCAA--AGAAGCUUUCAAAG-CGGCUAUA-ACAAUGGCGGCAAGCUGAUGAAAAAAAAA ----((((((((((((((.......)).)))))))))))).(((((..--....)))))....(-(.(((((.-...))))).)).................. ( -28.00, z-score = -2.00, R) >droSec1.super_13 1400834 95 + 2104621 ----GAUUUGGGUUAAGAUUUUUCCUCUUUGGCCCAGAUCAAAGACAA--AGAAGCUUUCAAAG-CGGCUAUA-ACAAUGGCGGCAAGCUGAUGAAAAAUAAA ----((((((((((((((.......)).))))))))))))........--...(((((.....(-(.(((((.-...))))).)))))))............. ( -27.60, z-score = -2.46, R) >droYak2.chr3R 4965774 92 - 28832112 ----GAUUUGGGUUAAGAUUUUUCCUCUGUGGCCCAGAUCAAAGGCAC--AGAAGCUUUCAAAG-CGGCUAUA-ACAAUGGCGGCAAACUGAAAAAAAAA--- ----((((((((((((((.......))).))))))))))).(((((..--....)))))....(-(.(((((.-...))))).))...............--- ( -28.00, z-score = -2.61, R) >droEre2.scaffold_4820 5053613 88 - 10470090 ----GAUUUGGGUUAAGAUUUUUCCUCUGUGGCCCAGAUCAAAGGCAA---AGAGCUUUCAAAG-CGGCUAUA-ACAAUGGCGGCAAACUGAAAAAA------ ----((((((((((((((.......))).))))))))))).(((((..---...)))))....(-(.(((((.-...))))).))............------ ( -27.80, z-score = -2.58, R) >droAna3.scaffold_13340 6991459 88 - 23697760 ----GAUUUGGGUUAAAAUUUUCCUCUGUUGGCCCAGAUCAAAGGCAAGAAGAAGCCCCCGAAGUCAGCUCCAUACAAUGGCCGC----UGAAAAA------- ----((((((((((((............))))))))))))...(((........))).......(((((.((((...))))..))----)))....------- ( -26.50, z-score = -2.11, R) >droWil1.scaffold_181130 13946925 84 - 16660200 ----CUUUUGGGUCAUCUUUUGUCCAGAUCAAAGUUAGUUAGCCAAAUAAUAGAAACUUCAAAAGAAGCGAAGCCUGGCCAUAAAACG--------------- ----.((((((((((..((((((........(((((.((((......))))...)))))........))))))..))))).)))))..--------------- ( -12.89, z-score = 0.11, R) >droMoj3.scaffold_6540 4317299 96 - 34148556 AUUAGAUUUGGGUCAG--UUUGUUUUUCGAAACGCAGCUUAGAUGAAA---GACAAGAACAACAACAGCUACG-CCAGCAACAAUGGGCCAACAGAAGAUAA- .......((((.(((.--(((((((((((...(........).)))))---))))))..........(((...-..))).....))).))))..........- ( -16.20, z-score = 0.03, R) >consensus ____GAUUUGGGUUAAGAUUUUUCCUCUUUGGCCCAGAUCAAAGGCAA__AGAAGCUUUCAAAG_CGGCUAUA_ACAAUGGCGGCAAGCUGAAAAAAAA____ ....((((((((((((((.......))).)))))))))))............................................................... ( -8.61 = -9.32 + 0.72)
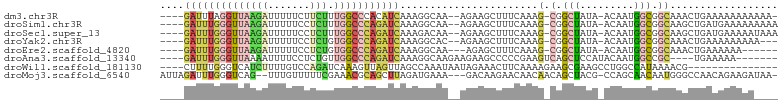


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:38 2011