| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,609,050 – 22,609,162 |
| Length | 112 |
| Max. P | 0.791494 |

| Location | 22,609,050 – 22,609,162 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.57 |
| Shannon entropy | 0.67761 |
| G+C content | 0.37438 |
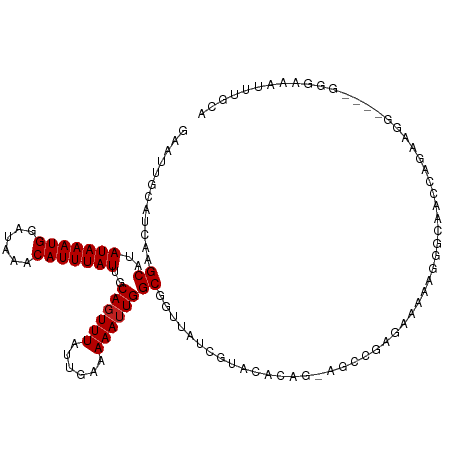
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -8.57 |
| Energy contribution | -8.65 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
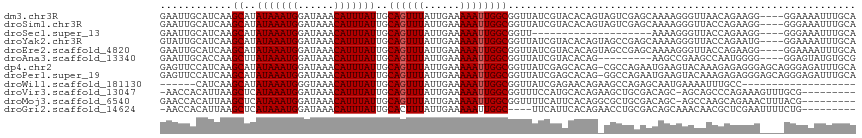
>dm3.chr3R 22609050 112 - 27905053 GAAUUGCAUCAAGCAUAUAAAUGGAUAAACAUUUAUUGCAGUUUAUUGAAAAAUUGGCGGUUAUCGUACACAGUAGUCGAGCAAAAGGGUUAACAGAAGG----GGAAAAUUUGCA .((((((.....(((.(((((((......))))))))))(((((......))))).)))))).(((.((......)))))(((((....((..(....).----.))...))))). ( -20.20, z-score = -1.14, R) >droSim1.chr3R 22440612 112 - 27517382 GAAUUGCAUCAAGCAUAUAAAUGGAUAAACAUUUAUUGCAGUUUAUUGAAAAAUUGGCGGUUAUCGUACACAGUAGUCGAGCAAAAGGGUUACCAGAAGG----GGGAAAUUUGCA .((((((.....(((.(((((((......))))))))))(((((......))))).)))))).(((.((......)))))(((((....(..((....))----..)...))))). ( -22.20, z-score = -1.25, R) >droSec1.super_13 1383261 92 - 2104621 GAAUUGCAUCAAGCAUAUAAAUGGAUAAACAUUUAUUGCAGUUUAUUGAAAAAUUGGCGGUU--------------------AAAAGGGUUACCAGAAGG----GGGAAAUUUGCA ....((((....(((.(((((((......)))))))))).((..(((....)))..))....--------------------....((....))......----........)))) ( -16.30, z-score = -1.00, R) >droYak2.chr3R 4947433 112 + 28832112 GUAUUGCAUCAAGCAUAUAAAUGGAUAAACAUUUAUUGCAGUUUAUUGAAAAAUUGGCGGUUAUCGUACACAGUAGCCGAGCAAAAGGGUUACCAGAAUG----GGAAAAUUUGCA (((((((.((..(((.(((((((......)))))))))).((..(((....)))..))((((((.(....).))))))))))))..((....))......----........))). ( -25.60, z-score = -1.66, R) >droEre2.scaffold_4820 5035974 112 + 10470090 GAAUUGCAUCAAGCAUAUAAAUGGAUAAACAUUUAUUGCAGUUUAUUGAAAAAUUGGCGGUUAUCGUACACAGUAGCCGAGCAAAAGGGUUACCAGAAGG----GGAAAAUUUGCA ((((((((........(((((((......))))))))))))))).............(((((((.(....).))))))).(((((....(..((....))----..)...))))). ( -25.00, z-score = -1.83, R) >droAna3.scaffold_13340 6972730 103 + 23697760 GAAUUGCACCAAGCUUAUAAAUGGAUAAACAUUUAUUGCAGUUUAUUGAAAAAUUGGCGGUUAUCGUACACAG---------AAGCCGAAGCCAAUGGGG----GGAGUAUGUGCG .....((((...((..(((((((......))))))).)).............((((((((((.((.......)---------)))))...))))))....----.......)))). ( -23.60, z-score = -0.65, R) >dp4.chr2 25924296 115 + 30794189 GAGUUCCAUCAAGCAUAUAAAUGGAUAAACAUUUAUUGCAGUUUAUUGAAAAAUUGGCGGUUAUCGAGCACAG-CGCCAGAAUGAAGUACAAAGAGAGGGAGCAGGGAGAUUUGCA ..(((((.((..(((.(((((((......))))))))))(.((((((......((((((.(..........).-)))))))))))).).......)).)))))............. ( -25.30, z-score = -1.52, R) >droPer1.super_19 1301611 115 + 1869541 GAGUUCCAUCAAGCAUAUAAAUGGAUAAACAUUUAUUGCAGUUUAUUGAAAAAUUGGCGGUUAUCGAGCACAG-GGCCAGAAUGAAGUACAAAGAGAGGGAGCAGGGAGAUUUGCA ..(((((.((..(((.(((((((......))))))))))(.((((((......(((((.(((....)))....-.))))))))))).).......)).)))))............. ( -23.50, z-score = -1.10, R) >droWil1.scaffold_181130 13910572 91 + 16660200 ------CAUCAAGCAUAUAAAUGGGUAAACAUUUAUUGCAGUUUAUUGAAAAAUUGGCGGUUAUCGAGAACAGAAGCCAGAGCAAUGAAAAUUUGCC------------------- ------.................((((((..((((((((.((..(((....)))..))((((.((.......))))))...))))))))..))))))------------------- ( -21.10, z-score = -2.44, R) >droVir3.scaffold_13047 11916048 105 - 19223366 -AACCACAUUAAGCUCAUAAAUGGAUAAACAUUUAUUGCAGUUUAUUGAAAAAUUGGCGGUUUCCAUGCACAGAAGCUGCGACAGC-AGCAGCCCAGAAAGUUUGCG--------- -.....((.((((((((((((((......)))))).)).)))))).)).......(((((...))..........(((((....))-))).))).............--------- ( -25.10, z-score = -1.11, R) >droMoj3.scaffold_6540 4285844 106 + 34148556 GAACCACAUUAAGCUCAUAAAUGGAUAAACAUUUAUUGCAGUUUAUUGAAAAAUUGGCGGUUUUCAUUCACAGGCGCUGCGACAGC-AGCCAAGCAGAAACUUUACG--------- (((((.......((..(((((((......))))))).)).((..(((....)))..)))))))..........(((((((....))-)))...))............--------- ( -24.20, z-score = -1.75, R) >droGri2.scaffold_14624 2053644 102 - 4233967 -AACCACAUUAAGCUCAUAAAUGGAUAAACAUUUAUUGCACUUUAUUGAAAAAUUGGC----UUCAUUCACAGAACCUGCGACAGCAAACAACGCUCGAAUUUUCUG--------- -...........((..(((((((......))))))).))........(((((.(((((----(((.......)))..(((....)))......)).))).)))))..--------- ( -15.80, z-score = -1.36, R) >consensus GAAUUGCAUCAAGCAUAUAAAUGGAUAAACAUUUAUUGCAGUUUAUUGAAAAAUUGGCGGUUAUCGUACACAG_AGCCGAGAAAAAGGGCAACCAGAAGG____GGGAAAUUUGCA ............((..(((((((......)))))))..((((((......)))))))).......................................................... ( -8.57 = -8.65 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:34 2011