
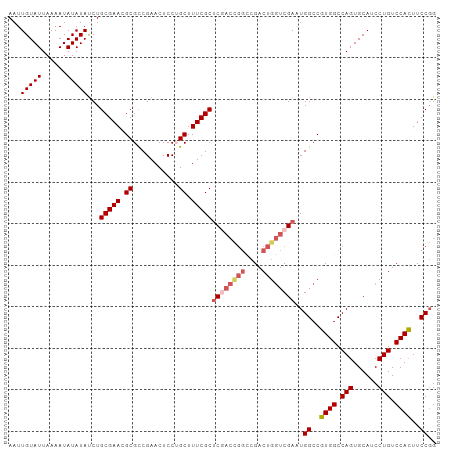
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,589,809 – 22,589,912 |
| Length | 103 |
| Max. P | 0.774754 |

| Location | 22,589,809 – 22,589,912 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 91.75 |
| Shannon entropy | 0.15066 |
| G+C content | 0.53245 |
| Mean single sequence MFE | -31.54 |
| Consensus MFE | -23.80 |
| Energy contribution | -25.12 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
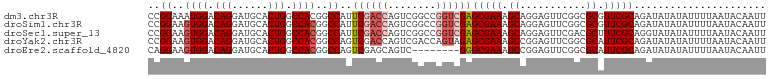
>dm3.chr3R 22589809 103 + 27905053 AAUUGUAUUAAAAUAUAUAUCUGCGAACGCGCCGAACUCCUGCUUUCGCUCGACCGGCCGACUGGUCGAAUGGCCGUGGCCAGUGCAUCCUGUCCAUUUCCGG ...(((((......)))))...(((((.(((.........))).)))))((((((((....)))))))).(((..((((.(((......))).))))..))). ( -30.00, z-score = -1.16, R) >droSim1.chr3R 22421105 103 + 27517382 AAUUGUAUUAAAAUAUAUAUCUGCGAACGCGCCGAACUCCUGCUUUCGCUCGACCGGCCGACUGGUCGAAUGGCCGUGGCCAGUGCAUCCUGUCCACUUCCGG ...(((((......)))))...(((((.(((.........))).)))))((((((((....)))))))).(((..((((.(((......))).))))..))). ( -31.50, z-score = -1.71, R) >droSec1.super_13 1363808 103 + 2104621 AAUUGUAUUAAAAUAUAUACCUGCGAAAGCGUCGAACUCCUGCUUUCGCUCGACCGGCCGACUGGUCGAAUGGCCGUGGCCAGUGCAUCCUGUCCACUUCCGG ....((((........))))..(((((((((.........)))))))))((((((((....)))))))).(((..((((.(((......))).))))..))). ( -35.80, z-score = -3.32, R) >droYak2.chr3R 4927510 103 - 28832112 AAUUGUAUUAAAAUAUAUAUCUGCGAAUGCGCCGAACUCCGGCUUUCGCUCUACUGGUCGACUGGUCGACUGGCCGUGGCCAGUGCAUCCUGUCCACUUCCGG ...(((((......)))))...(((((...((((.....)))).)))))....((((..(((.((..(((((((....)))))).)..)).))).....)))) ( -33.90, z-score = -2.79, R) >droEre2.scaffold_4820 5013050 95 - 10470090 AAUUGUAUUAAAAUAUAUAUCUGCGAAUGCGCCGAACUCCGGCUUUCGCCC--------GACUGCUCGACUGGCCGUGGCCAGUGCAUCCUGUCCACUUCCUG ...(((((......)))))...(((((...((((.....)))).)))))..--------((((((...((((((....)))))))))....)))......... ( -26.50, z-score = -2.35, R) >consensus AAUUGUAUUAAAAUAUAUAUCUGCGAACGCGCCGAACUCCUGCUUUCGCUCGACCGGCCGACUGGUCGAAUGGCCGUGGCCAGUGCAUCCUGUCCACUUCCGG ...(((((......)))))...(((((.((...........)).)))))((((((((....))))))))..((..((((.(((......))).))))..)).. (-23.80 = -25.12 + 1.32)


| Location | 22,589,809 – 22,589,912 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 91.75 |
| Shannon entropy | 0.15066 |
| G+C content | 0.53245 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -24.28 |
| Energy contribution | -25.52 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.694552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22589809 103 - 27905053 CCGGAAAUGGACAGGAUGCACUGGCCACGGCCAUUCGACCAGUCGGCCGGUCGAGCGAAAGCAGGAGUUCGGCGCGUUCGCAGAUAUAUAUUUUAAUACAAUU (((....)))...((((((.(((..(.(((((..((((....))))..))))).((....))....)..))).))))))........................ ( -31.00, z-score = -0.73, R) >droSim1.chr3R 22421105 103 - 27517382 CCGGAAGUGGACAGGAUGCACUGGCCACGGCCAUUCGACCAGUCGGCCGGUCGAGCGAAAGCAGGAGUUCGGCGCGUUCGCAGAUAUAUAUUUUAAUACAAUU (((((.((((.(((......))).))))..((..((((((.(....).))))))((....)).))..))))).((....))...................... ( -33.10, z-score = -1.19, R) >droSec1.super_13 1363808 103 - 2104621 CCGGAAGUGGACAGGAUGCACUGGCCACGGCCAUUCGACCAGUCGGCCGGUCGAGCGAAAGCAGGAGUUCGACGCUUUCGCAGGUAUAUAUUUUAAUACAAUU ..((..((((.(((......))).))))..))..((((((.(....).))))))((((((((.(........)))))))))..((((........)))).... ( -37.60, z-score = -2.95, R) >droYak2.chr3R 4927510 103 + 28832112 CCGGAAGUGGACAGGAUGCACUGGCCACGGCCAGUCGACCAGUCGACCAGUAGAGCGAAAGCCGGAGUUCGGCGCAUUCGCAGAUAUAUAUUUUAAUACAAUU ..((..((((.(((......))).))))..)).(((((....))))).......(((((.((((.....))))...)))))...................... ( -33.90, z-score = -2.69, R) >droEre2.scaffold_4820 5013050 95 + 10470090 CAGGAAGUGGACAGGAUGCACUGGCCACGGCCAGUCGAGCAGUC--------GGGCGAAAGCCGGAGUUCGGCGCAUUCGCAGAUAUAUAUUUUAAUACAAUU ..((..((((.(((......))).))))..)).(((((((..((--------.(((....))).)))))))))((....))...................... ( -36.00, z-score = -4.08, R) >consensus CCGGAAGUGGACAGGAUGCACUGGCCACGGCCAUUCGACCAGUCGGCCGGUCGAGCGAAAGCAGGAGUUCGGCGCAUUCGCAGAUAUAUAUUUUAAUACAAUU ..((..((((.(((......))).))))..))..((((((........))))))(((((.((...........)).)))))...................... (-24.28 = -25.52 + 1.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:31 2011