| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,570,350 – 22,570,454 |
| Length | 104 |
| Max. P | 0.999065 |

| Location | 22,570,350 – 22,570,454 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.86 |
| Shannon entropy | 0.35991 |
| G+C content | 0.42795 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -23.84 |
| Energy contribution | -23.86 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.995184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
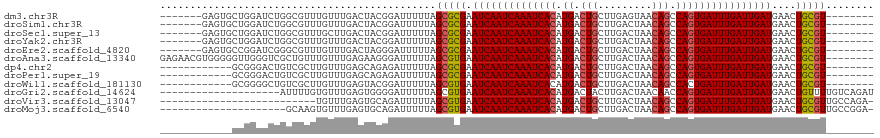
>dm3.chr3R 22570350 104 + 27905053 -------GAGUGCUGGAUCUGGCGUUUGUUUGACUACGGAUUUUUAGCGCGAAUCAAUCAAAUCACAUGACUGCUUGAGUAACAGCCAGUGAUUUGAUUGAUGAACUGCGU-------- -------..((((((((((((..(((.....)))..))))))..))))))..((((((((((((((.((.(((.........))).)))))))))))))))).........-------- ( -36.20, z-score = -3.58, R) >droSim1.chr3R 22401672 104 + 27517382 -------GAGUGCUGGAUCUGGCGUUUGUUUGACUACGGAUUUUUAGCGCGAAUCAAUCAAAUCACAUGACUGCUUGACUAACAGCCAGUGAUUUGAUUGAUGAACUGCGU-------- -------..((((((((((((..(((.....)))..))))))..))))))..((((((((((((((.((.(((.........))).)))))))))))))))).........-------- ( -36.20, z-score = -4.00, R) >droSec1.super_13 1344486 104 + 2104621 -------GAGUGCUGGAUCUGGCGUUUGCUUGACUACGGAUUUUUAGCGCGAAUCAAUCAAAUCACAUGACUGCUUGACUAACAGCCAGUGAUUUGAUUGAUGAACUGCGU-------- -------..((((((((((((..(((.....)))..))))))..))))))..((((((((((((((.((.(((.........))).)))))))))))))))).........-------- ( -36.20, z-score = -3.99, R) >droYak2.chr3R 4906768 104 - 28832112 -------GAGUGCUGGAUCUGGCGUUUGUUUGACUACGGAUUUUUAGCGCGAAUCAAUCAAAUCACAUGACUGCUUGACUAACAGCCAGUGAUUUGAUUGAUGAACUGCGU-------- -------..((((((((((((..(((.....)))..))))))..))))))..((((((((((((((.((.(((.........))).)))))))))))))))).........-------- ( -36.20, z-score = -4.00, R) >droEre2.scaffold_4820 4993482 104 - 10470090 -------GAGUGCCGGAUCGGGCGUUUGUUUGACUAGGGAUUUUUAGCGCGAAUCAAUCAAAUCACAUGACUGCUUGACUAACAGCCAGUGAUUUGAUUGAUGAACUGCGU-------- -------((((.((...((((((....))))))...)).))))...(((((.((((((((((((((.((.(((.........))).))))))))))))))))....)))))-------- ( -34.90, z-score = -3.03, R) >droAna3.scaffold_13340 6934969 111 - 23697760 GAGAACGUGGGGGUUGGGUCGCUGUUUGUUUGAGAAGGGAUUUUUAGCGUGAAUCAAUCAAAUCACAUGACUGCUUGACUAACAGCCAGUGAUUUGAUUGAUGAACUGCGU-------- ....(((..(..((((((...((.(((......))).))...))))))....((((((((((((((.((.(((.........))).))))))))))))))))...)..)))-------- ( -33.10, z-score = -1.96, R) >dp4.chr2 25881761 99 - 30794189 ------------GCGGGACUGUCGCUUGUUUGAGCAGAGAUUUUUAGCGCGAAUCAAUCAAAUCACAUGACUGCUUGACUAACAGCCAGUGAUUUGAUUGAUGAACUGCGU-------- ------------((((...((.((((.((((......))))....)))))).((((((((((((((.((.(((.........))).))))))))))))))))...))))..-------- ( -31.50, z-score = -2.32, R) >droPer1.super_19 1259222 99 - 1869541 ------------GCGGGACUGUCGCUUGUUUGAGCAGAGAUUUUUAGCGCGAAUCAAUCAAAUCACAUGACUGCUUGACUAACAGCCAGUGAUUUGAUUGAUGAACUGCGU-------- ------------((((...((.((((.((((......))))....)))))).((((((((((((((.((.(((.........))).))))))))))))))))...))))..-------- ( -31.50, z-score = -2.32, R) >droWil1.scaffold_181130 13832151 99 - 16660200 ------------GCGGGGCUGUCGCUUGUUUGAGUACGGAUUUUUAGCGUGAAUCAAUCAAAUCACAUGACUGCUUGACUAACAGCCACUGAUUUGAUUGAUGAACUGCGU-------- ------------((((..(...((((.(((((....)))))....))))...(((((((((((((..((.(((.........))).)).))))))))))))))..))))..-------- ( -29.40, z-score = -2.10, R) >droGri2.scaffold_14624 1984968 99 + 4233967 --------------------AUUUUGUGUUUGAGUGGGGAUUUUUAGCGUGAAUCAAUCAAAUCACAUGACUACUUGACUAACAACCAGUGAUUUGAUUGAUGAACUGUUUUGUCAGAU --------------------..................(((....(((((..((((((((((((((.((.................))))))))))))))))..)).)))..))).... ( -22.23, z-score = -0.69, R) >droVir3.scaffold_13047 11865170 92 + 19223366 --------------------------UGUUUGAGUGCAGAUUUUUAGCGUGAAUCAAUCAAAUCACAUGACUGCUUGACUAACAGCCAGUGAUUUGAUUGAUGAACUGCGUUGCCAGA- --------------------------.......(.((((.......((((..((((((((((((((.((.(((.........))).))))))))))))))))..)).)).)))))...- ( -27.80, z-score = -2.64, R) >droMoj3.scaffold_6540 4237555 97 - 34148556 ---------------------GCAAGUGUUUGAGUGCAGAUUUUUAGCGUGAAUCAAUCAAAUCACAUGACUGCUUGACUAACAGCCAGUGAUUUGAUUGAUGAACUGCGUUGCCGGA- ---------------------((((..(((((....))))).....((((..((((((((((((((.((.(((.........))).))))))))))))))))..)).)).))))....- ( -30.00, z-score = -2.29, R) >consensus ____________CUGGAUCUGGCGUUUGUUUGAGUACGGAUUUUUAGCGCGAAUCAAUCAAAUCACAUGACUGCUUGACUAACAGCCAGUGAUUUGAUUGAUGAACUGCGU________ ..............................................(((((.((((((((((((((.((.(((.........))).))))))))))))))))....)))))........ (-23.84 = -23.86 + 0.01)

| Location | 22,570,350 – 22,570,454 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.86 |
| Shannon entropy | 0.35991 |
| G+C content | 0.42795 |
| Mean single sequence MFE | -32.41 |
| Consensus MFE | -29.94 |
| Energy contribution | -29.72 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.84 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
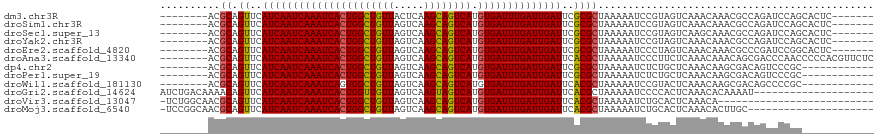
>dm3.chr3R 22570350 104 - 27905053 --------ACGCAGUUCAUCAAUCAAAUCACUGGCUGUUACUCAAGCAGUCAUGUGAUUUGAUUGAUUCGCGCUAAAAAUCCGUAGUCAAACAAACGCCAGAUCCAGCACUC------- --------..((.((..((((((((((((((((((((((.....)))))))).))))))))))))))..))((((........))))...................))....------- ( -33.10, z-score = -4.99, R) >droSim1.chr3R 22401672 104 - 27517382 --------ACGCAGUUCAUCAAUCAAAUCACUGGCUGUUAGUCAAGCAGUCAUGUGAUUUGAUUGAUUCGCGCUAAAAAUCCGUAGUCAAACAAACGCCAGAUCCAGCACUC------- --------..((.((..((((((((((((((((((((((.....)))))))).))))))))))))))..))((((........))))...................))....------- ( -33.10, z-score = -4.88, R) >droSec1.super_13 1344486 104 - 2104621 --------ACGCAGUUCAUCAAUCAAAUCACUGGCUGUUAGUCAAGCAGUCAUGUGAUUUGAUUGAUUCGCGCUAAAAAUCCGUAGUCAAGCAAACGCCAGAUCCAGCACUC------- --------..((.((..((((((((((((((((((((((.....)))))))).))))))))))))))..))..............(((..((....))..)))...))....------- ( -34.40, z-score = -4.90, R) >droYak2.chr3R 4906768 104 + 28832112 --------ACGCAGUUCAUCAAUCAAAUCACUGGCUGUUAGUCAAGCAGUCAUGUGAUUUGAUUGAUUCGCGCUAAAAAUCCGUAGUCAAACAAACGCCAGAUCCAGCACUC------- --------..((.((..((((((((((((((((((((((.....)))))))).))))))))))))))..))((((........))))...................))....------- ( -33.10, z-score = -4.88, R) >droEre2.scaffold_4820 4993482 104 + 10470090 --------ACGCAGUUCAUCAAUCAAAUCACUGGCUGUUAGUCAAGCAGUCAUGUGAUUUGAUUGAUUCGCGCUAAAAAUCCCUAGUCAAACAAACGCCCGAUCCGGCACUC------- --------..((.((..((((((((((((((((((((((.....)))))))).))))))))))))))..)))).......................(((......)))....------- ( -35.70, z-score = -5.68, R) >droAna3.scaffold_13340 6934969 111 + 23697760 --------ACGCAGUUCAUCAAUCAAAUCACUGGCUGUUAGUCAAGCAGUCAUGUGAUUUGAUUGAUUCACGCUAAAAAUCCCUUCUCAAACAAACAGCGACCCAACCCCCACGUUCUC --------..((.((..((((((((((((((((((((((.....)))))))).))))))))))))))..)))).............................................. ( -31.20, z-score = -5.52, R) >dp4.chr2 25881761 99 + 30794189 --------ACGCAGUUCAUCAAUCAAAUCACUGGCUGUUAGUCAAGCAGUCAUGUGAUUUGAUUGAUUCGCGCUAAAAAUCUCUGCUCAAACAAGCGACAGUCCCGC------------ --------.(((.((..((((((((((((((((((((((.....)))))))).))))))))))))))..))((...........))........)))..........------------ ( -34.90, z-score = -4.85, R) >droPer1.super_19 1259222 99 + 1869541 --------ACGCAGUUCAUCAAUCAAAUCACUGGCUGUUAGUCAAGCAGUCAUGUGAUUUGAUUGAUUCGCGCUAAAAAUCUCUGCUCAAACAAGCGACAGUCCCGC------------ --------.(((.((..((((((((((((((((((((((.....)))))))).))))))))))))))..))((...........))........)))..........------------ ( -34.90, z-score = -4.85, R) >droWil1.scaffold_181130 13832151 99 + 16660200 --------ACGCAGUUCAUCAAUCAAAUCAGUGGCUGUUAGUCAAGCAGUCAUGUGAUUUGAUUGAUUCACGCUAAAAAUCCGUACUCAAACAAGCGACAGCCCCGC------------ --------.(((.((..((((((((((((((((((((((.....))))))))).)))))))))))))..))...........((......))..)))..........------------ ( -31.30, z-score = -4.46, R) >droGri2.scaffold_14624 1984968 99 - 4233967 AUCUGACAAAACAGUUCAUCAAUCAAAUCACUGGUUGUUAGUCAAGUAGUCAUGUGAUUUGAUUGAUUCACGCUAAAAAUCCCCACUCAAACACAAAAU-------------------- .............((..((((((((((((((((..((((.....))))..)).))))))))))))))..))............................-------------------- ( -20.70, z-score = -2.44, R) >droVir3.scaffold_13047 11865170 92 - 19223366 -UCUGGCAACGCAGUUCAUCAAUCAAAUCACUGGCUGUUAGUCAAGCAGUCAUGUGAUUUGAUUGAUUCACGCUAAAAAUCUGCACUCAAACA-------------------------- -...(....)((((...((((((((((((((((((((((.....)))))))).)))))))))))))).............)))).........-------------------------- ( -32.89, z-score = -5.45, R) >droMoj3.scaffold_6540 4237555 97 + 34148556 -UCCGGCAACGCAGUUCAUCAAUCAAAUCACUGGCUGUUAGUCAAGCAGUCAUGUGAUUUGAUUGAUUCACGCUAAAAAUCUGCACUCAAACACUUGC--------------------- -...(....)(((((..((((((((((((((((((((((.....)))))))).))))))))))))))..))((.........))...........)))--------------------- ( -33.60, z-score = -5.13, R) >consensus ________ACGCAGUUCAUCAAUCAAAUCACUGGCUGUUAGUCAAGCAGUCAUGUGAUUUGAUUGAUUCGCGCUAAAAAUCCGUACUCAAACAAACGCCAGAUCCAG____________ ..........((.((..((((((((((((((((((((((.....)))))))).))))))))))))))..)))).............................................. (-29.94 = -29.72 + -0.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:30 2011