| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,511,674 – 22,511,786 |
| Length | 112 |
| Max. P | 0.629412 |

| Location | 22,511,674 – 22,511,786 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 73.19 |
| Shannon entropy | 0.49910 |
| G+C content | 0.41044 |
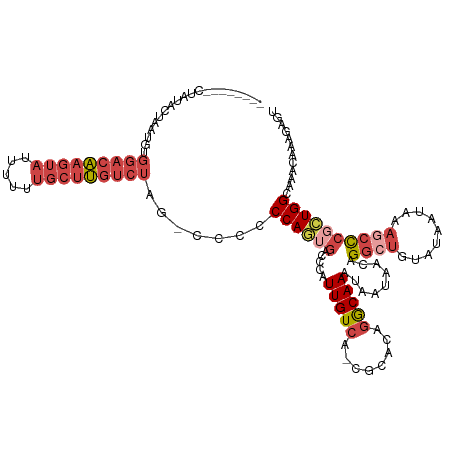
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -16.86 |
| Energy contribution | -18.83 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22511674 112 - 27905053 --------CUAUACUAAUGUGGACAAGUAUUUUUUGCUUGUCUUG--CCCCCAGUGACCCAUUGUCA-CGCACAGGCAAAUAAUAACAGGCUGUAUAAUAAAGCCCGCUGGCAAACAAAGAGU --------.(((((...(((.((((((((.....))))))))(((--((....(((((.....))))-).....)))))......)))....))))).....(((....)))........... ( -31.00, z-score = -1.92, R) >droEre2.scaffold_4820 4929826 112 + 10470090 --------CUGUACUAAUGUGGACAAGUAUUUUUUGCUUGUCUA--UUCCCCAGUGACCCGUUGUCA-CGCACAGGCAAAUAAUAACAGGCUGAAUAAUAAAGCCCGCUGGCAAACAAAGCGU --------((((......(((((((((((.....))))))))))--)......(((((.....))))-)..)))).............((((.........))))((((.........)))). ( -31.20, z-score = -1.90, R) >droYak2.chr3R 4846693 112 + 28832112 --------CUAUACAAAUGUGGACAAGUAUUUUUUGCUUGUCUA--UUCCCCAGUGACCCAUUGUCA-CGCACAGGCAAAUAAUAACAGGCUGAAUACAAAAACUCGCUGGCAAACAAAGAGU --------.........((((((((((((.....)))))))))(--(((.((.(((((.....))))-)((....))...........))..)))))))...((((..((.....))..)))) ( -26.80, z-score = -1.66, R) >droSec1.super_13 1285818 122 - 2104621 CUGUACAACUAUACUAAUGUGGACAAGUAUUUUUUGCUUGUCUUGUCCCCCCAGUGACCCAUUGUCA-CGCACAGGCAAAUAAUAACAGGCUGUAUAAUAAAGCCCGAUGGCAAACAAAGAGU .(((.....(((((...(((.((((((((.....))))))))(((((......(((((.....))))-).....)))))......)))....))))).....(((....)))..)))...... ( -28.70, z-score = -0.91, R) >droSim1.chr3R 22340780 122 - 27517382 CUAUACAACUAUACUAAUGUGGACAAGUAUUUUUUGCUUGUCUUGUCCCCCCAGAGACCCAUUGUCA-CGCACAGGCAAAUAAUAACAGGCUGUAUAAUAAAGCCCGCUGGCAAACAAAGAGU ............(((..((((((((((((.....))))))))).......((((.(.....(((((.-......))))).........((((.........))))).))))...)))...))) ( -25.30, z-score = -0.18, R) >droWil1.scaffold_181130 9982690 92 + 16660200 ----------------------CGGCGUAUUAUUAUAUCGUACAUACAUGCCAAAUAUCAGUUGUAAUUGCGCAAACAAAU-GUAAUGGG--------UAAACGUUGUUGGCGAAAAGCAAAA ----------------------..(((((((((......))).)))).((((((......(((...((((((........)-)))))...--------..)))....))))))....)).... ( -15.20, z-score = 1.37, R) >consensus ________CUAUACUAAUGUGGACAAGUAUUUUUUGCUUGUCUAG_CCCCCCAGUGACCCAUUGUCA_CGCACAGGCAAAUAAUAACAGGCUGUAUAAUAAAGCCCGCUGGCAAACAAAGAGU ....................(((((((((.....))))))))).......((((((.....(((((........))))).........((((.........))))))))))............ (-16.86 = -18.83 + 1.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:26 2011