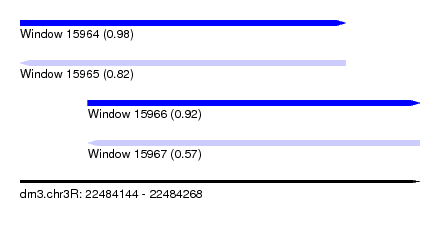
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,484,144 – 22,484,268 |
| Length | 124 |
| Max. P | 0.980160 |

| Location | 22,484,144 – 22,484,245 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.85 |
| Shannon entropy | 0.31592 |
| G+C content | 0.45109 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -18.04 |
| Energy contribution | -18.42 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
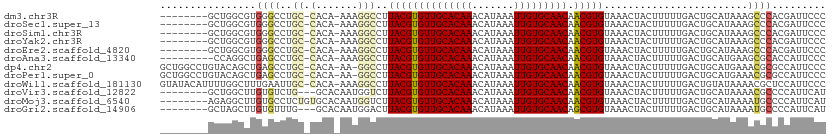
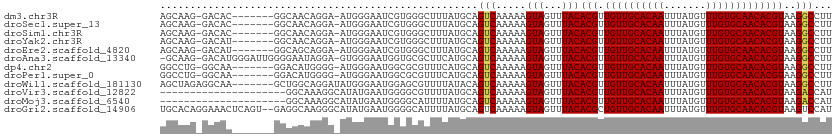
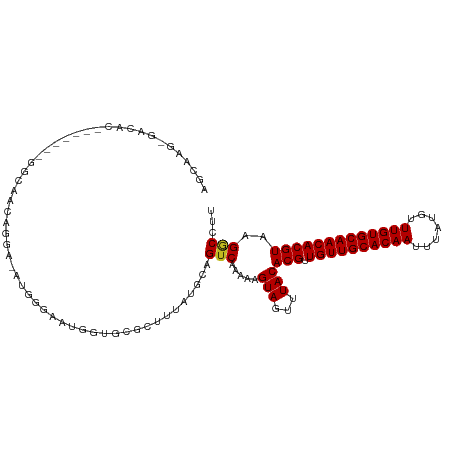
>dm3.chr3R 22484144 101 + 27905053 --------GCUGGCGUGGGCCUGC-CACA-AAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAGCCCACGAUUCCC --------.....(((((((.(((-..((-(((((...((((((((((((((.......))))))))).))))).......)))))))...)))....)))))))...... ( -36.60, z-score = -4.06, R) >droSec1.super_13 1258544 101 + 2104621 --------GCUGGCGUGGGCCUGC-CACA-AAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAGCCCACGAUUCCC --------.....(((((((.(((-..((-(((((...((((((((((((((.......))))))))).))))).......)))))))...)))....)))))))...... ( -36.60, z-score = -4.06, R) >droSim1.chr3R 22312789 101 + 27517382 --------GCUGGCGUGGGCCUGC-CACA-AAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAGCCCACGAUUCCC --------.....(((((((.(((-..((-(((((...((((((((((((((.......))))))))).))))).......)))))))...)))....)))))))...... ( -36.60, z-score = -4.06, R) >droYak2.chr3R 4818405 101 - 28832112 --------GCUGGCGUGGGCCUGC-CACA-AAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAGCCCACGAUUCCC --------.....(((((((.(((-..((-(((((...((((((((((((((.......))))))))).))))).......)))))))...)))....)))))))...... ( -36.60, z-score = -4.06, R) >droEre2.scaffold_4820 4900561 101 - 10470090 --------GCUGGCGUGGGCCUGC-CACA-AAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAGCCCACGAUUCCC --------.....(((((((.(((-..((-(((((...((((((((((((((.......))))))))).))))).......)))))))...)))....)))))))...... ( -36.60, z-score = -4.06, R) >droAna3.scaffold_13340 6846466 100 - 23697760 ---------CCAGGCUGAGCCUGC-CACA-AAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUGAAGCGCACCAUUCCC ---------.(((((...))))).-..((-(((((...((((((((((((((.......))))))))).))))).......)))))))..(((.......)))........ ( -28.10, z-score = -2.05, R) >dp4.chr2 10499758 108 + 30794189 GCUGGCCUGUACAGCUGAGCCUGC-CACA-AA-GGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUGAAACGCGCCAUUCCC ..((((.(((.((((.(.((((..-....-.)-)))..((((((((((((((.......))))))))).)))))...............).)).))..))).))))..... ( -30.70, z-score = -1.76, R) >droPer1.super_0 1841659 108 - 11822988 GCUGGCCUGUACAGCUGAGCCUGC-CACA-AA-GGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUGAAACGCGCCAUUCCC ..((((.(((.((((.(.((((..-....-.)-)))..((((((((((((((.......))))))))).)))))...............).)).))..))).))))..... ( -30.70, z-score = -1.76, R) >droWil1.scaffold_181130 9949151 109 - 16660200 GUAUACAUUUUGGCUUUGAAUUGC-CACA-AAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGUAUAAAACGCUCCAUUCCC .((((((...((((........))-))((-(((((...((((((((((((((.......))))))))).))))).......)))))))..))))))............... ( -30.50, z-score = -3.94, R) >droVir3.scaffold_12822 648756 100 - 4096053 --------GCUGGCUUGUGUCUG---GCACAAUGGUCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAACGCCCCAUUCAU --------...((((((((....---.)))))((((..((((((((((((((.......))))))))).)))))...))))...................)))........ ( -27.50, z-score = -1.87, R) >droMoj3.scaffold_6540 17392172 103 - 34148556 --------AGAGGCUUGUGCCUCUGUGCACAAUGGUCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAAUGCCCCAUUCAU --------.(((((....)))))(((((((((((((..((((((((((((((.......))))))))).)))))...))))....)))..))))))............... ( -33.20, z-score = -3.59, R) >droGri2.scaffold_14906 13101367 100 - 14172833 --------GCUAGCUUGUGUUUG---GCACAAUGGACUUACGUGUUGCACAAACAUAAAUUGUGCAACAGCGUGUAAACUACUUUUUGACUGCAUAAAAUGCCCCAUUCAU --------((((((....).)))---))..(((((...((((((((((((((.......)))))))))).)))).................((((...)))).)))))... ( -25.90, z-score = -1.09, R) >consensus ________GCUGGCGUGGGCCUGC_CACA_AAAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAACGCACCAUUCCC ................((((.(((...((....((...((((((((((((((.......))))))))).)))))....))......))...)))....))))......... (-18.04 = -18.42 + 0.38)
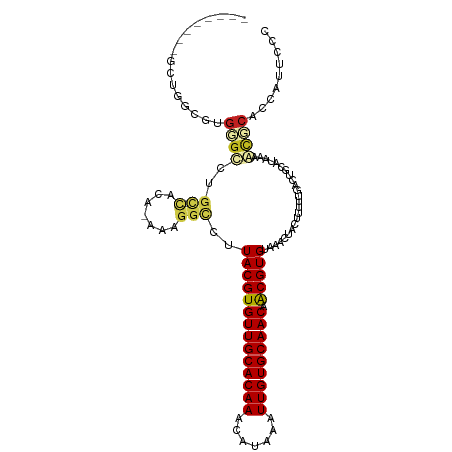
| Location | 22,484,144 – 22,484,245 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.85 |
| Shannon entropy | 0.31592 |
| G+C content | 0.45109 |
| Mean single sequence MFE | -34.79 |
| Consensus MFE | -19.93 |
| Energy contribution | -19.53 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.823626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22484144 101 - 27905053 GGGAAUCGUGGGCUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUUU-UGUG-GCAGGCCCACGCCAGC-------- .((...((((((((...(((...(((((.((.......((((.(((((((((.......)))))))))))))...)).)))-))..-)))))))))))))...-------- ( -39.00, z-score = -3.38, R) >droSec1.super_13 1258544 101 - 2104621 GGGAAUCGUGGGCUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUUU-UGUG-GCAGGCCCACGCCAGC-------- .((...((((((((...(((...(((((.((.......((((.(((((((((.......)))))))))))))...)).)))-))..-)))))))))))))...-------- ( -39.00, z-score = -3.38, R) >droSim1.chr3R 22312789 101 - 27517382 GGGAAUCGUGGGCUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUUU-UGUG-GCAGGCCCACGCCAGC-------- .((...((((((((...(((...(((((.((.......((((.(((((((((.......)))))))))))))...)).)))-))..-)))))))))))))...-------- ( -39.00, z-score = -3.38, R) >droYak2.chr3R 4818405 101 + 28832112 GGGAAUCGUGGGCUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUUU-UGUG-GCAGGCCCACGCCAGC-------- .((...((((((((...(((...(((((.((.......((((.(((((((((.......)))))))))))))...)).)))-))..-)))))))))))))...-------- ( -39.00, z-score = -3.38, R) >droEre2.scaffold_4820 4900561 101 + 10470090 GGGAAUCGUGGGCUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUUU-UGUG-GCAGGCCCACGCCAGC-------- .((...((((((((...(((...(((((.((.......((((.(((((((((.......)))))))))))))...)).)))-))..-)))))))))))))...-------- ( -39.00, z-score = -3.38, R) >droAna3.scaffold_13340 6846466 100 + 23697760 GGGAAUGGUGCGCUUCAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUUU-UGUG-GCAGGCUCAGCCUGG--------- (((....(.((((.....)).(((((((((..((((..((((.(((((((((.......))))))))))))).))))))))-).))-))..)).)..)))..--------- ( -30.60, z-score = -0.71, R) >dp4.chr2 10499758 108 - 30794189 GGGAAUGGCGCGUUUCAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCC-UU-UGUG-GCAGGCUCAGCUGUACAGGCCAGC .....((((((((...)))).)))).............((((.(((((((((.......)))))))))))))..((((-(.-((..-((.......))..)).)))))... ( -37.40, z-score = -1.85, R) >droPer1.super_0 1841659 108 + 11822988 GGGAAUGGCGCGUUUCAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCC-UU-UGUG-GCAGGCUCAGCUGUACAGGCCAGC .....((((((((...)))).)))).............((((.(((((((((.......)))))))))))))..((((-(.-((..-((.......))..)).)))))... ( -37.40, z-score = -1.85, R) >droWil1.scaffold_181130 9949151 109 + 16660200 GGGAAUGGAGCGUUUUAUACAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUUU-UGUG-GCAAUUCAAAGCCAAAAUGUAUAC ...............((((((....(((((..((((..((((.(((((((((.......))))))))))))).))))))))-).((-((........))))...)))))). ( -28.00, z-score = -1.47, R) >droVir3.scaffold_12822 648756 100 + 4096053 AUGAAUGGGGCGUUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGACCAUUGUGC---CAGACACAAGCCAGC-------- ........(((((((((((..........(((...)))....((((((((((.......))))))))))))))))))..(((((.---....))))))))...-------- ( -28.20, z-score = -1.20, R) >droMoj3.scaffold_6540 17392172 103 + 34148556 AUGAAUGGGGCAUUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGACCAUUGUGCACAGAGGCACAAGCCUCU-------- ......(((((..........(((.....(((...)))((((.(((((((((.......)))))))))))))..)))..((((((......))))))))))).-------- ( -34.90, z-score = -3.18, R) >droGri2.scaffold_14906 13101367 100 + 14172833 AUGAAUGGGGCAUUUUAUGCAGUCAAAAAGUAGUUUACACGCUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGUCCAUUGUGC---CAAACACAAGCUAGC-------- .......(((((((((.((....)).))))).......(((.((((((((((.......)))))))))))))..)))).(((((.---....)))))......-------- ( -26.00, z-score = -0.54, R) >consensus GGGAAUGGUGCGCUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUUU_UGUG_GCAGGCCCACGCCAGC________ .........(.((((..(((((((.....(((...)))(((.((((((((((.......)))))))))))))..))).....))))...)))).)................ (-19.93 = -19.53 + -0.40)

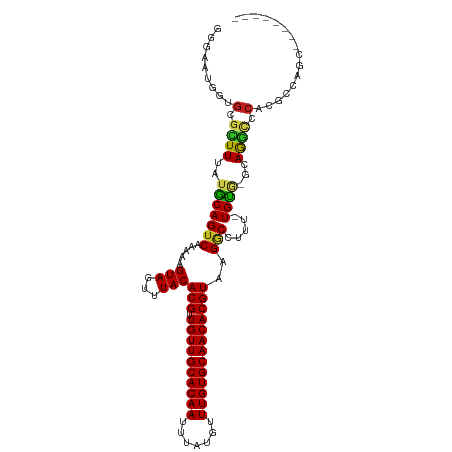
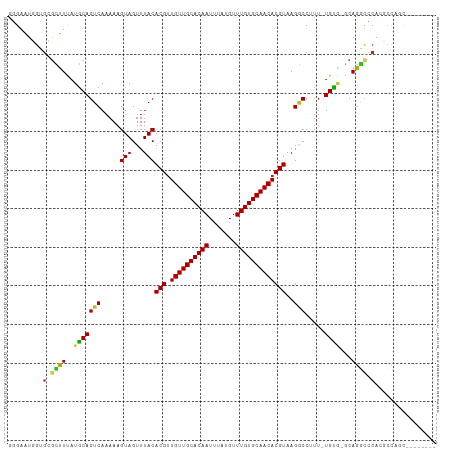
| Location | 22,484,165 – 22,484,268 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.77 |
| Shannon entropy | 0.36675 |
| G+C content | 0.43277 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -16.74 |
| Energy contribution | -16.67 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22484165 103 + 27905053 AAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAGCCCACGAUUCCCAU-UCCUGUUGCC-------GUGUC-CUUGCU .((....((((((((((((((.......))))))))).)))))....))...........(((...((..((((........-.........)-------)))..-))))). ( -23.53, z-score = -1.61, R) >droSec1.super_13 1258565 103 + 2104621 AAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAGCCCACGAUUCCCAU-UCCUGUUGCC-------GUGUC-CUUGCU .((....((((((((((((((.......))))))))).)))))....))...........(((...((..((((........-.........)-------)))..-))))). ( -23.53, z-score = -1.61, R) >droSim1.chr3R 22312810 103 + 27517382 AAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAGCCCACGAUUCCCAU-UCCUGUUGCC-------GUGUC-CUUGCU .((....((((((((((((((.......))))))))).)))))....))...........(((...((..((((........-.........)-------)))..-))))). ( -23.53, z-score = -1.61, R) >droYak2.chr3R 4818426 103 - 28832112 AAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAGCCCACGAUUCCCAU-UCCUGUUGCC-------AUGUC-CUUGCU ..(((..((((((((((((((.......))))))))).))))).......((((.((....)).))))..............-.......)))-------.....-...... ( -22.60, z-score = -1.52, R) >droEre2.scaffold_4820 4900582 103 - 10470090 AAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAGCCCACGAUUCCCAU-UCCUGCUGCC-------AUGUC-CUUGCU ..(((..((((((((((((((.......))))))))).))))).......((((.((....)).))))..............-.......)))-------.....-...... ( -22.60, z-score = -1.55, R) >droAna3.scaffold_13340 6846486 109 - 23697760 AAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUGAAGCGCACCAUUCCCAC-UCCUAUUCCCCAAUCCCAUGUC-CUUGC- ...(((((...((((((((((.......)))))))))).((((((..(........)..))))))))).))...........-......................-.....- ( -20.90, z-score = -1.54, R) >dp4.chr2 10499786 103 + 30794189 AAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUGAAACGCGCCAUUCCCAU-CCCCAUGUCC-------UUGCC-CAGGCC ..(((((((((((((((((((.......))))))))).))))).................(((((.................-...)))))..-------.....-.))))) ( -26.15, z-score = -2.17, R) >droPer1.super_0 1841687 103 - 11822988 AAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUGAAACGCGCCAUUCCCAU-CCCCAUGUCC-------UUGCC-CAGGCC ..(((((((((((((((((((.......))))))))).))))).................(((((.................-...)))))..-------.....-.))))) ( -26.15, z-score = -2.17, R) >droWil1.scaffold_181130 9949180 105 - 16660200 AAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGUAUAAAACGCUCCAUUCCCAUAUCCUGCCAGC-------UUGCCUCUAGCU ..(((..((((((((((((((.......))))))))).))))).............((.(((.....))).))..............)))(((-------(.......)))) ( -23.80, z-score = -2.38, R) >droVir3.scaffold_12822 648776 91 - 4096053 AUGGUCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAACGCCCCAUUCAUAUGCCUUUGCC--------------------- .((((..((((((((((((((.......))))))))).)))))...))))..........(((((..............))))).......--------------------- ( -21.14, z-score = -2.17, R) >droMoj3.scaffold_6540 17392195 91 - 34148556 AUGGUCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAAUGCCCCAUUCAUAUGCCUUUGCC--------------------- .((((..((((((((((((((.......))))))))).)))))...))))..........(((((.((((...))))..))))).......--------------------- ( -22.50, z-score = -2.64, R) >droGri2.scaffold_14906 13101387 110 - 14172833 AUGGACUUACGUGUUGCACAAACAUAAAUUGUGCAACAGCGUGUAAACUACUUUUUGACUGCAUAAAAUGCCCCAUUCAUAUGCCCUUGCCUC--ACUGAGUUUCCUGUGCA ..(((..((((((((((((((.......)))))))))).))))......((((..(((..(((((.((((...))))..))))).......))--)..)))).)))...... ( -24.60, z-score = -1.14, R) >consensus AAGGCCUUACGUGUUGCACAAACAUAAAUUGUGCAACAACGUGUAAACUACUUUUUGACUGCAUAAAACGCACCAUUCCCAU_UCCUGUUGCC_______GUGUC_CUUGCU .......((((((((((((((.......))))))))).)))))..................................................................... (-16.74 = -16.67 + -0.08)

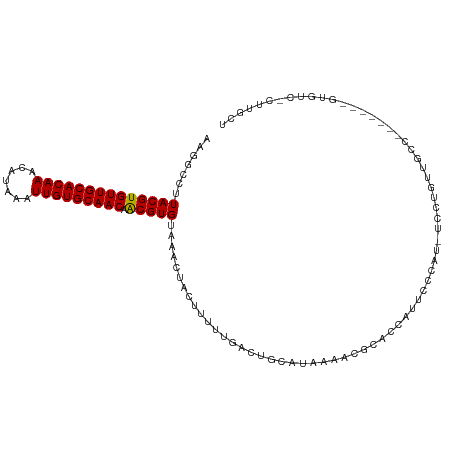
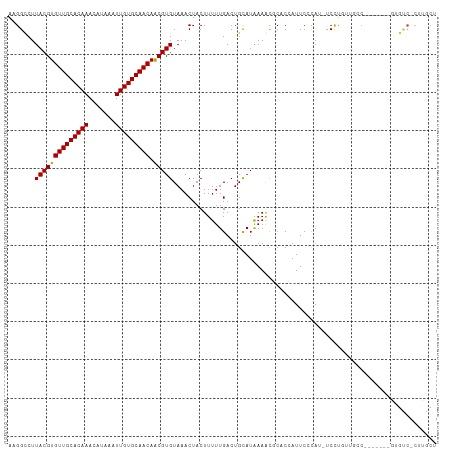
| Location | 22,484,165 – 22,484,268 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.77 |
| Shannon entropy | 0.36675 |
| G+C content | 0.43277 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -18.01 |
| Energy contribution | -17.97 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.569617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22484165 103 - 27905053 AGCAAG-GACAC-------GGCAACAGGA-AUGGGAAUCGUGGGCUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUU ......-..(((-------((...((...-.))....)))))(((((((((..........(((...)))....((((((((((.......))))))))))))))))))).. ( -29.80, z-score = -1.75, R) >droSec1.super_13 1258565 103 - 2104621 AGCAAG-GACAC-------GGCAACAGGA-AUGGGAAUCGUGGGCUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUU ......-..(((-------((...((...-.))....)))))(((((((((..........(((...)))....((((((((((.......))))))))))))))))))).. ( -29.80, z-score = -1.75, R) >droSim1.chr3R 22312810 103 - 27517382 AGCAAG-GACAC-------GGCAACAGGA-AUGGGAAUCGUGGGCUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUU ......-..(((-------((...((...-.))....)))))(((((((((..........(((...)))....((((((((((.......))))))))))))))))))).. ( -29.80, z-score = -1.75, R) >droYak2.chr3R 4818426 103 + 28832112 AGCAAG-GACAU-------GGCAACAGGA-AUGGGAAUCGUGGGCUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUU ......-..(((-------((...((...-.))....)))))(((((((((..........(((...)))....((((((((((.......))))))))))))))))))).. ( -28.00, z-score = -1.16, R) >droEre2.scaffold_4820 4900582 103 + 10470090 AGCAAG-GACAU-------GGCAGCAGGA-AUGGGAAUCGUGGGCUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUU ......-..(((-------((...((...-.))....)))))(((((((((..........(((...)))....((((((((((.......))))))))))))))))))).. ( -26.90, z-score = -0.52, R) >droAna3.scaffold_13340 6846486 109 + 23697760 -GCAAG-GACAUGGGAUUGGGGAAUAGGA-GUGGGAAUGGUGCGCUUCAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUU -..(((-(.(....(((((.(.....(((-(((.(.....).)))))).).)))))..............((((.(((((((((.......)))))))))))))..).)))) ( -29.80, z-score = -1.10, R) >dp4.chr2 10499786 103 - 30794189 GGCCUG-GGCAA-------GGACAUGGGG-AUGGGAAUGGCGCGUUUCAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUU (((((.-(((..-------.(.(((.(..-(((.(.....).)))..))))).)))..............((((.(((((((((.......))))))))))))).))))).. ( -34.80, z-score = -2.26, R) >droPer1.super_0 1841687 103 + 11822988 GGCCUG-GGCAA-------GGACAUGGGG-AUGGGAAUGGCGCGUUUCAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUU (((((.-(((..-------.(.(((.(..-(((.(.....).)))..))))).)))..............((((.(((((((((.......))))))))))))).))))).. ( -34.80, z-score = -2.26, R) >droWil1.scaffold_181130 9949180 105 + 16660200 AGCUAGAGGCAA-------GCUGGCAGGAUAUGGGAAUGGAGCGUUUUAUACAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUU .....(((((((-------((((....(((...((((((...)))))).....)))......))))))..((((.(((((((((.......)))))))))))))...))))) ( -28.20, z-score = -1.12, R) >droVir3.scaffold_12822 648776 91 + 4096053 ---------------------GGCAAAGGCAUAUGAAUGGGGCGUUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGACCAU ---------------------(((....(((((.(((((...)))))))))).)))........((((..((((.(((((((((.......))))))))))))).))))... ( -25.10, z-score = -1.71, R) >droMoj3.scaffold_6540 17392195 91 + 34148556 ---------------------GGCAAAGGCAUAUGAAUGGGGCAUUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGACCAU ---------------------(((....(((((.(((((...)))))))))).)))........((((..((((.(((((((((.......))))))))))))).))))... ( -25.50, z-score = -2.10, R) >droGri2.scaffold_14906 13101387 110 + 14172833 UGCACAGGAAACUCAGU--GAGGCAAGGGCAUAUGAAUGGGGCAUUUUAUGCAGUCAAAAAGUAGUUUACACGCUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGUCCAU ..(((((....))..))--)......((((...(((.....((((...))))..))).............(((.((((((((((.......)))))))))))))..)))).. ( -30.70, z-score = -1.42, R) >consensus AGCAAG_GACAC_______GGCAACAGGA_AUGGGAAUGGUGCGCUUUAUGCAGUCAAAAAGUAGUUUACACGUUGUUGCACAAUUUAUGUUUGUGCAACACGUAAGGCCUU .....................................................(((.....(((...)))(((.((((((((((.......)))))))))))))..)))... (-18.01 = -17.97 + -0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:23 2011