| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,451,190 – 22,451,245 |
| Length | 55 |
| Max. P | 0.647961 |

| Location | 22,451,190 – 22,451,245 |
|---|---|
| Length | 55 |
| Sequences | 12 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 77.02 |
| Shannon entropy | 0.51389 |
| G+C content | 0.58942 |
| Mean single sequence MFE | -19.24 |
| Consensus MFE | -9.23 |
| Energy contribution | -9.17 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22451190 55 + 27905053 ---GCUGGUAGUUGUAGCCGAGCUUAUCGGCGCAGGCGACUGCCACCAGGCACAUUAC ---((((((((((((.(((((.....)))))....)))))))))....)))....... ( -24.60, z-score = -3.10, R) >droSim1.chr3R 22279181 55 + 27517382 ---GCUGGUAGUUGUAGCCGAGCUUAUCGGCGCAGGUGACUGCCACCAGGCACAUUAC ---.(((((.......(((((.....)))))((((....)))).)))))......... ( -24.50, z-score = -3.06, R) >droSec1.super_13 1226019 55 + 2104621 ---GCUGGUAGUUGUAGCCGAGCUUAUCGGCGCAGGUGACUGCCACCAGGCACAUUAC ---.(((((.......(((((.....)))))((((....)))).)))))......... ( -24.50, z-score = -3.06, R) >droYak2.chr3R 24038582 52 + 28832112 ---GCUGGUAGUUGUAGCCA---CUCCUGGCACUGACGGCAGCCACGAGGCACAGAGC ---((((.((((....((((---....)))))))).)))).(((....)))....... ( -16.60, z-score = 0.24, R) >droEre2.scaffold_4820 4867563 55 - 10470090 ---GCUGGUAGUUGUAGCCGAGCUUAUCGGCGCAGGCGGCUGCCACCAGGCACAGUAC ---((((((((((((.(((((.....)))))....)))))))))....)))....... ( -24.10, z-score = -1.76, R) >droAna3.scaffold_13340 6812905 55 - 23697760 ---GCUGGUAAUUGUAGCCAAGUUUGUCGGCGCAGGCGACGGCCACCAGGCACAUAAC ---.(((((...(((.(((.........))))))(((....))))))))......... ( -16.20, z-score = -0.28, R) >dp4.chr2 22313568 52 - 30794189 GACGGUUGUAGUUGUAGCCGG---CCUCAGGACGGGCCGCGGCCA-GGGCCAGACA-- ...(((((......)))))((---(((......)))))..(((..-..))).....-- ( -19.20, z-score = 0.01, R) >droPer1.super_0 10009726 55 + 11822988 ---GCUUGUAGUUGUAGCCGAGUUUGUCGGCGCAGGCGACGGCCACGAGGCACAGGAC ---.(((((.(((((.(((((.....)))))....))))).(((....)))))))).. ( -22.10, z-score = -1.90, R) >droWil1.scaffold_181130 9913946 55 - 16660200 ---GCUGGUAGUUGUAUCCCAAUUUAUCGGCGCAGGCAACUGCCACCAGGCAUAUAAC ---(((((((((((.....))).))))))))((((....))))............... ( -17.40, z-score = -1.86, R) >droVir3.scaffold_12822 3481500 55 + 4096053 ---GCUGGUAGUUGUAGCCAAGUUUAUCGGCACAGGCAACGGCCACAAGGCACAAGAC ---((((((.(((((.(((.........)))....))))).)))....)))....... ( -14.40, z-score = -0.92, R) >droMoj3.scaffold_6540 17356137 55 - 34148556 ---GCUGGUAGUUAUAGCCGAGCUUAUCGGCGCACGCAACUGCGAUCAGGCACACAAC ---.(((((.......(((((.....)))))...(((....))))))))......... ( -17.10, z-score = -1.47, R) >droGri2.scaffold_14906 1099228 51 + 14172833 ---GCUUAUAGUUGUAACCCAGUUUAUCGGCACAGGCAACGGCCACAAGGCACA---- ---.......(((((...((........)).....))))).(((....)))...---- ( -10.20, z-score = -0.13, R) >consensus ___GCUGGUAGUUGUAGCCGAGCUUAUCGGCGCAGGCGACUGCCACCAGGCACAUAAC ...((((((.((.((.(((...............))).)).)).)))).))....... ( -9.23 = -9.17 + -0.06)

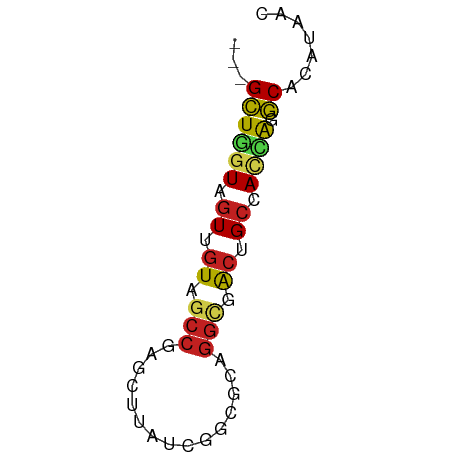
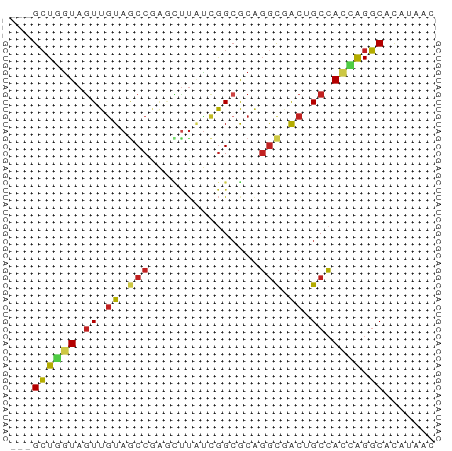
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:15 2011