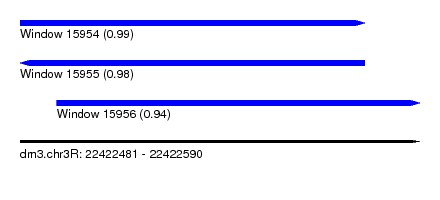
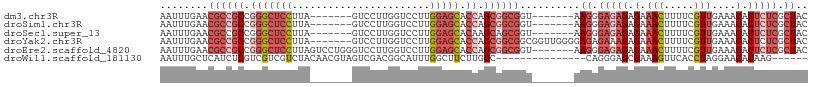
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,422,481 – 22,422,590 |
| Length | 109 |
| Max. P | 0.993006 |

| Location | 22,422,481 – 22,422,575 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 74.40 |
| Shannon entropy | 0.46995 |
| G+C content | 0.53684 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -21.45 |
| Energy contribution | -24.12 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.993006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
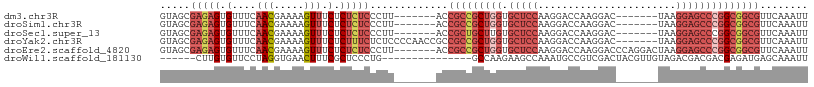
>dm3.chr3R 22422481 94 + 27905053 GUAGCGAGAGUGUUUCAACGAAAAGUUUCUCUCUCCCUU-------ACCGCCGCUGGUGCUCCAAGGACCAAGGAC-------UAAGGAGCCCGGCGGCGUUCAAAUU ((((.(((((.(....(((.....))).).))))).).)-------))(((((((((.(((((..((.(....).)-------)..))))))))))))))........ ( -37.70, z-score = -3.30, R) >droSim1.chr3R 22250798 94 + 27517382 GUAGCGAGAGUGUUUCAACGAAAAGUUUCUCUCUCCCUU-------ACCGCCGCUGGUGCUCCAAGGACCAAGGAC-------UAAGGAGCCCGGCGGCGUUCAAAUU ((((.(((((.(....(((.....))).).))))).).)-------))(((((((((.(((((..((.(....).)-------)..))))))))))))))........ ( -37.70, z-score = -3.30, R) >droSec1.super_13 1197481 94 + 2104621 GUAGCGAGAGUGUUUCAACGAAAAGUUUCUCUCUCCCUU-------ACCGCUGCUUGUGCUCCAAGGACCAAGGAC-------UAAGGAGCCCGGCGGCGUUCAAAUU ((((.(((((.(....(((.....))).).))))).).)-------))(((((((.(.(((((..((.(....).)-------)..)))))).)))))))........ ( -29.70, z-score = -1.24, R) >droYak2.chr3R 4754209 101 - 28832112 GUAGCGAGAGUGUUUCAACGAAAAGUUUCUCUUUCUCUCCCCAACCGCCGCCGCUGGUGCUCCAAGGACCAAGGAC-------UAAGGAGCCCGGCGGCGUUCAAAUU .....(((((......(((.....))).......))))).........(((((((((.(((((..((.(....).)-------)..))))))))))))))........ ( -36.42, z-score = -2.25, R) >droEre2.scaffold_4820 4836716 101 - 10470090 GUAGCGAGAGUGUUUCAACGAAAAGUUUCUCUCUCCCUU-------ACCGCCGCUGGUGCUCCAAGGACCAAGGACCCAGGACUAAGGAGCCCGGCGGCGUUCAAAUU ((((.(((((.(....(((.....))).).))))).).)-------))(((((((((.(((((..((.((.........)).))..))))))))))))))........ ( -38.80, z-score = -2.68, R) >droWil1.scaffold_181130 9881714 87 - 16660200 ------CUUGUGUUCCUAGGUGAACUUUCGCUCCCUG---------------GCCAAGAAGCCAAAUGCCGUCGACUACGUUGUAGACGACGACGAGAUGAGCAAAUU ------.....((((......))))....((((..((---------------((......))))..((.(((((.((((...)))).))))).))....))))..... ( -23.10, z-score = -1.17, R) >consensus GUAGCGAGAGUGUUUCAACGAAAAGUUUCUCUCUCCCUU_______ACCGCCGCUGGUGCUCCAAGGACCAAGGAC_______UAAGGAGCCCGGCGGCGUUCAAAUU .....(((((.(....(((.....))).).))))).............(((((((((.(((((.......................))))))))))))))........ (-21.45 = -24.12 + 2.67)
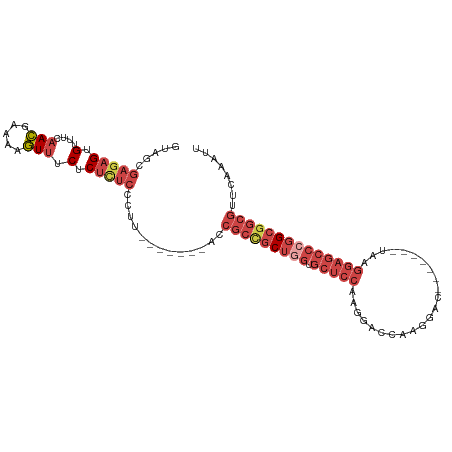
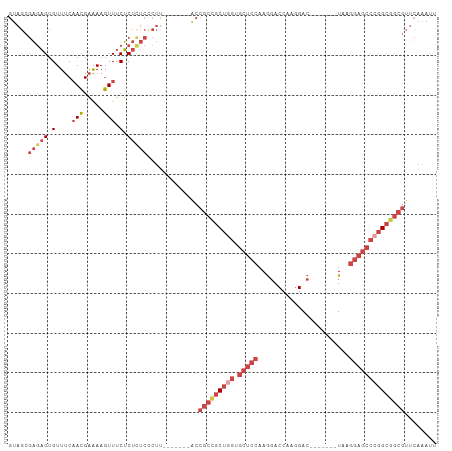
| Location | 22,422,481 – 22,422,575 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 74.40 |
| Shannon entropy | 0.46995 |
| G+C content | 0.53684 |
| Mean single sequence MFE | -35.55 |
| Consensus MFE | -19.53 |
| Energy contribution | -22.18 |
| Covariance contribution | 2.65 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22422481 94 - 27905053 AAUUUGAACGCCGCCGGGCUCCUUA-------GUCCUUGGUCCUUGGAGCACCAGCGGCGGU-------AAGGGAGAGAGAAACUUUUCGUUGAAACACUCUCGCUAC ........((((((.(((((((...-------(.(....).)...))))).)).))))))..-------.((.(((((.(((....)))((....)).))))).)).. ( -36.10, z-score = -2.26, R) >droSim1.chr3R 22250798 94 - 27517382 AAUUUGAACGCCGCCGGGCUCCUUA-------GUCCUUGGUCCUUGGAGCACCAGCGGCGGU-------AAGGGAGAGAGAAACUUUUCGUUGAAACACUCUCGCUAC ........((((((.(((((((...-------(.(....).)...))))).)).))))))..-------.((.(((((.(((....)))((....)).))))).)).. ( -36.10, z-score = -2.26, R) >droSec1.super_13 1197481 94 - 2104621 AAUUUGAACGCCGCCGGGCUCCUUA-------GUCCUUGGUCCUUGGAGCACAAGCAGCGGU-------AAGGGAGAGAGAAACUUUUCGUUGAAACACUCUCGCUAC ........(((.((..((((((...-------(.(....).)...))))).)..)).)))..-------.((.(((((.(((....)))((....)).))))).)).. ( -26.90, z-score = 0.04, R) >droYak2.chr3R 4754209 101 + 28832112 AAUUUGAACGCCGCCGGGCUCCUUA-------GUCCUUGGUCCUUGGAGCACCAGCGGCGGCGGUUGGGGAGAGAAAGAGAAACUUUUCGUUGAAACACUCUCGCUAC ........((((((((((((((...-------(.(....).)...))))).))...)))))))..(((.(((((...((((....))))((....)).))))).))). ( -38.30, z-score = -1.54, R) >droEre2.scaffold_4820 4836716 101 + 10470090 AAUUUGAACGCCGCCGGGCUCCUUAGUCCUGGGUCCUUGGUCCUUGGAGCACCAGCGGCGGU-------AAGGGAGAGAGAAACUUUUCGUUGAAACACUCUCGCUAC ........((((((.(((((((..((..(..(....)..)..)).))))).)).))))))..-------.((.(((((.(((....)))((....)).))))).)).. ( -39.50, z-score = -1.94, R) >droWil1.scaffold_181130 9881714 87 + 16660200 AAUUUGCUCAUCUCGUCGUCGUCUACAACGUAGUCGACGGCAUUUGGCUUCUUGGC---------------CAGGGAGCGAAAGUUCACCUAGGAACACAAG------ ..(((((((.....(((((((.((((...)))).))))))).(((((((....)))---------------))))))))))).((((......)))).....------ ( -36.40, z-score = -5.37, R) >consensus AAUUUGAACGCCGCCGGGCUCCUUA_______GUCCUUGGUCCUUGGAGCACCAGCGGCGGU_______AAGGGAGAGAGAAACUUUUCGUUGAAACACUCUCGCUAC ........((((((.(((((((.......................))))).)).))))))..........((.(((((.(.(((.....)))....).))))).)).. (-19.53 = -22.18 + 2.65)
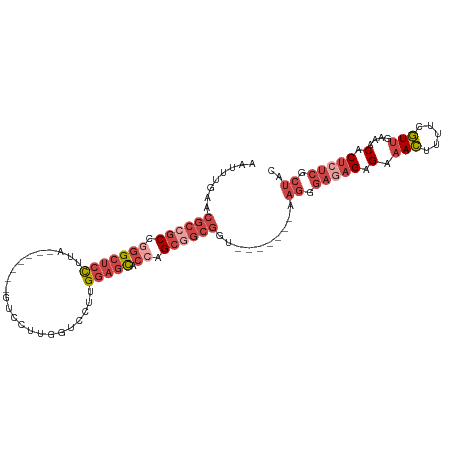
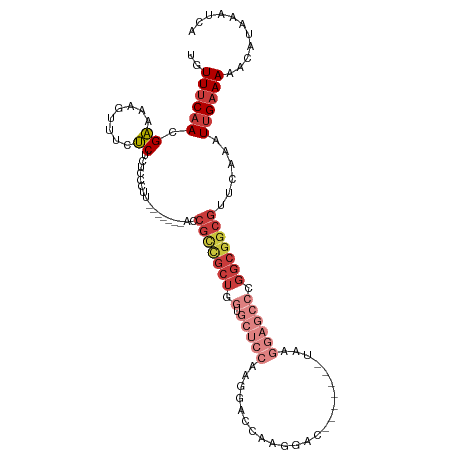
| Location | 22,422,491 – 22,422,590 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.50 |
| Shannon entropy | 0.50003 |
| G+C content | 0.46831 |
| Mean single sequence MFE | -28.29 |
| Consensus MFE | -11.59 |
| Energy contribution | -13.87 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22422491 99 + 27905053 UGUUUCAACGAAAAGUUUCUCUCUCCCUU-------ACCGCCGCUGGUGCUCCAAGGACCAAGGAC-------UAAGGAGCCCGGCGGCGUUCAAAUUGAAAAACAUAAAUCA ..((((((.(((....)))..........-------..(((((((((.(((((..((.(....).)-------)..))))))))))))))......))))))........... ( -32.80, z-score = -3.22, R) >droSim1.chr3R 22250808 99 + 27517382 UGUUUCAACGAAAAGUUUCUCUCUCCCUU-------ACCGCCGCUGGUGCUCCAAGGACCAAGGAC-------UAAGGAGCCCGGCGGCGUUCAAAUUGAAAAACAUAAAUCA ..((((((.(((....)))..........-------..(((((((((.(((((..((.(....).)-------)..))))))))))))))......))))))........... ( -32.80, z-score = -3.22, R) >droSec1.super_13 1197491 99 + 2104621 UGUUUCAACGAAAAGUUUCUCUCUCCCUU-------ACCGCUGCUUGUGCUCCAAGGACCAAGGAC-------UAAGGAGCCCGGCGGCGUUCAAAUUGAAAAACAUAAAUCA ..((((((.(((....)))..........-------..(((((((.(.(((((..((.(....).)-------)..)))))).)))))))......))))))........... ( -24.80, z-score = -1.11, R) >droYak2.chr3R 4754219 106 - 28832112 UGUUUCAACGAAAAGUUUCUCUUUCUCUCCCCAACCGCCGCCGCUGGUGCUCCAAGGACCAAGGAC-------UAAGGAGCCCGGCGGCGUUCAAAUUGAAAAACAUAAAUCA ..((((((.((((((.....)))).))...........(((((((((.(((((..((.(....).)-------)..))))))))))))))......))))))........... ( -34.10, z-score = -2.90, R) >droEre2.scaffold_4820 4836726 106 - 10470090 UGUUUCAACGAAAAGUUUCUCUCUCCCUU-------ACCGCCGCUGGUGCUCCAAGGACCAAGGACCCAGGACUAAGGAGCCCGGCGGCGUUCAAAUUGAAAAACAUAAAUCA ..((((((.(((....)))..........-------..(((((((((.(((((..((.((.........)).))..))))))))))))))......))))))........... ( -33.90, z-score = -2.48, R) >droAna3.scaffold_13340 6779446 78 - 23697760 UGUUUCAACGAAAAGUUUCUCU-UCUUUU--------CCGUUGCUCCU--UUUAACG------------------------CCGCCGGCGUUCAAAUUGAAAAACAUAAAUCA (((((((((((((((.......-.)))))--------.))))).((..--(((((((------------------------((...)))))).)))..)).)))))....... ( -16.50, z-score = -2.77, R) >droWil1.scaffold_181130 9881718 98 - 16660200 UGUUCCUAGGUGAACUUUCGCU--CCCUG-------------GCCAAGAAGCCAAAUGCCGUCGACUACGUUGUAGACGACGACGAGAUGAGCAAAUUGAAAAACAUAAAUCA .((((......))))....(((--(..((-------------((......))))..((.(((((.((((...)))).))))).))....)))).................... ( -23.10, z-score = -1.27, R) >consensus UGUUUCAACGAAAAGUUUCUCUCUCCCUU_______ACCGCCGCUGGUGCUCCAAGGACCAAGGAC_______UAAGGAGCCCGGCGGCGUUCAAAUUGAAAAACAUAAAUCA ..((((((.((........)).................(((((((.(.(((((.......................)))))).)))))))......))))))........... (-11.59 = -13.87 + 2.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:13 2011