
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,406,113 – 22,406,214 |
| Length | 101 |
| Max. P | 0.742685 |

| Location | 22,406,113 – 22,406,214 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 62.20 |
| Shannon entropy | 0.79131 |
| G+C content | 0.64887 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -15.58 |
| Energy contribution | -15.99 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.10 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22406113 101 + 27905053 CCUGGCAGCUGCUGCUGCCAUGGUGCAUGGCGCCUCGUCGCCCACACCACCUGCUGC---CACCUCCAAUAUAACUCCCUUACCCACAGCCGCGGCAGUAGCAU ..(((((((....)))))))(((((...((((......))))..))))).(((((((---(................................))))))))... ( -35.85, z-score = -1.84, R) >droGri2.scaffold_14906 13018744 89 - 14172833 CUUAGCAGCUGCGGCUGCCUUGGUGCAUGGCCCACCCUCGCCCUCCCCCAAACCAAC------------CACACCGAUUCCGUCCUCUGGAUCCGCGAUGC--- ....(((..(((((.((..((((.(...(((........)))...).))))..)).)------------).....(..((((.....))))..)))).)))--- ( -19.30, z-score = 0.88, R) >droMoj3.scaffold_6540 17305884 83 - 34148556 CUUAGCAGCAGCCGCUGCCCUGGUGCAUGGG------UCGCCAUCGCCCAUACGUAC------------AACACCAACGCCACCCGCUGGCUCGAUGAUGA--- ....(((((....)))))....(((((((((------.((....)))))))..))))------------.........((((.....))))..........--- ( -23.60, z-score = 0.75, R) >droVir3.scaffold_12822 557433 83 - 4096053 CCUGGCAGCAGCUGCUGCUCUUGUGCACGGG------GCGCCUUCACCCACACCCAC------------CACACCGACGCCGCCCGCUGGGUCUGCGGCAC--- ...((((((....))))))...((((..(((------.........)))........------------........(((.((((...))))..)))))))--- ( -26.50, z-score = 1.66, R) >droWil1.scaffold_181130 9863266 98 - 16660200 CUUGGCUGUUGCUGCUGCCUUGGUGCAUGGAACACCAUCUCCCACACCCACACCGACA------CCAAUCACACCAACACCGCCAGUAGAAGUACCACCACCAG ..(((.((.((((.((((..(((((.((((....))))................((..------....)).......)))))...)))).)))).))))).... ( -18.40, z-score = 0.48, R) >droAna3.scaffold_13340 6761190 86 - 23697760 CCUGGCAGCUGCUGCUGCCCUGGUGCACGGAGCCGCCUCACCGACGCCCACGAGUAC------ACCCAUUCCGACCCCCGUGCCAGUGGCAU------------ ...((((((....)))))).(((((..((....))...)))))..(((.((..((((------................))))..)))))..------------ ( -27.49, z-score = 0.23, R) >droEre2.scaffold_4820 4820285 101 - 10470090 UCUGGCAGCCGCUGCUGCCAUGGUGCACGGCGCUGCGUCGCCCACACCUCCUGCUGC---CACCUCCAAUACAACUCCAUUACCCACAGCCGCGCCAGUGGCAU ..(((((((.((....))...((((...((((......))))..))))....)))))---))..........................(((((....))))).. ( -33.40, z-score = -0.59, R) >droYak2.chr3R 4737535 104 - 28832112 CCUGGCAGCUGCUGCUGCCAUGGUGCAUGGCGCUUCGUCGCCCACACCACCUGCUGCUGCCACCUCCAAUACUACUCCCCAACCCGCAGCCGCUUCAGUGGCAU ..(((((((....((.....(((((...((((......))))..)))))...)).)))))))..........................(((((....))))).. ( -34.90, z-score = -1.08, R) >droSec1.super_13 1181234 101 + 2104621 CCUGGCAGCUGCUGCUGCCAUGGUGCAUGGCGCCUCGUCGCCCACACCACCUGCUGC---CACCUCCAAUACAACUCCCUUACCCACAGCCGCGCCAGUGGCAU ..(((((((.((....))..(((((...((((......))))..)))))...)))))---))..........................(((((....))))).. ( -34.70, z-score = -1.41, R) >droSim1.chr3R 22234078 101 + 27517382 CCUGGCAAUUGCUGCUGCCAUGGUGCAUGGCGCCUCGUCGCCCACACCACCUGCUGC---CACCUCCAAUACAACUCCCUUACCCACAGCCGCGCCAGUGGCAU ..(((((...((....))..(((((...((((......))))..))))).....)))---))..........................(((((....))))).. ( -28.40, z-score = -0.02, R) >consensus CCUGGCAGCUGCUGCUGCCAUGGUGCAUGGCGCCUCGUCGCCCACACCCACUGCUAC______CUCCAAUACAACUCCCCUACCCGCAGCCGCGCCAGUGGCAU ...((((((....))))))..((((...(((........)))..))))........................................................ (-15.58 = -15.99 + 0.41)

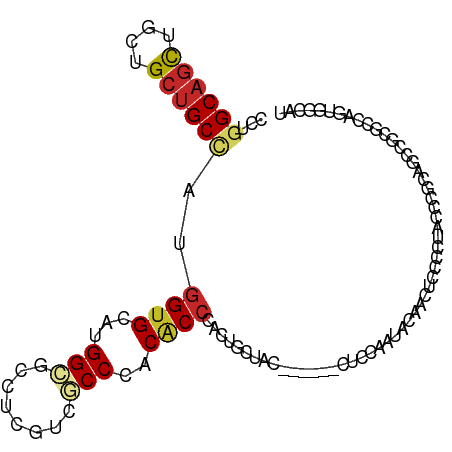

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:11 2011