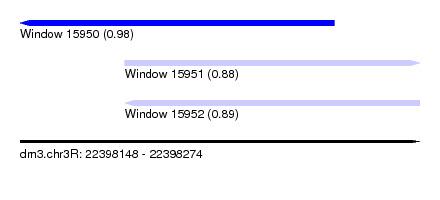
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,398,148 – 22,398,274 |
| Length | 126 |
| Max. P | 0.981475 |

| Location | 22,398,148 – 22,398,247 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 88.24 |
| Shannon entropy | 0.22165 |
| G+C content | 0.36409 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -20.51 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
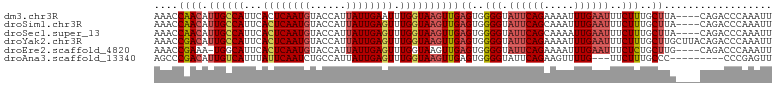
>dm3.chr3R 22398148 99 - 27905053 AAACCAACAUUGCCAUUCACUCAAUGUACCAUUAUUGAAUUUGGUAAGUUGAGUGGGGUAUUCAGAAAAUUUGAAUUUCUUUGCUUA----CAGACCCAAAUU .................((((((((.(((((..........))))).))))))))(((((((((((...)))))))....(((....----)))))))..... ( -23.60, z-score = -1.96, R) >droSim1.chr3R 22226093 99 - 27517382 AAACCAACAUUGCCAUUCACUCAAUGUACCAUUAUUGAGUUUGGUAAGUUGAGUGGGGUAUUCAGCAAAUUUGAAUUUCUUUGCUUA----CAGACCCAAAUU ..((((((.((((((...((((((((......)))))))).)))))))))).)).((((....((((((..........))))))..----...))))..... ( -30.10, z-score = -3.81, R) >droSec1.super_13 1173287 99 - 2104621 AAACCAACAUUGCCAUUCACUCAAUGUACCAUUAUUGAGUUUGGUAAGUUGAGUGGGGUAUUCAGCAAAAUUGAAUUUCUUUGCUUA----CAGACCCAAAUU ..((((((.((((((...((((((((......)))))))).)))))))))).)).((((....((((((..........))))))..----...))))..... ( -30.30, z-score = -3.86, R) >droYak2.chr3R 4729425 103 + 28832112 AAACCGACAUUGCCAUUCACUCAAUGUACCAUUAUUGAGUUUGGUAAGUUGAGUGGGGUAUUCAGAAAAUUUGAAUUUCUUUGCUUGCUUACAGACCCAAAUU ....((((.((((((...((((((((......)))))))).))))))))))((..(((.(((((((...)))))))..)))..)).................. ( -27.30, z-score = -2.51, R) >droEre2.scaffold_4820 4812412 98 + 10470090 AAACCGAAA-UGGCAUUCACUCAAUGUACCAUUAUUGAGUUUGGUAAGUUGAGUGGGGUAUUCAGAAAAUUUGAAUUUCUCUGCUUG----CAGACCCAAAUU .........-.......((((((((.(((((..........))))).))))))))(((((((((((...)))))))....(((....----)))))))..... ( -27.10, z-score = -2.37, R) >droAna3.scaffold_13340 6753296 91 + 23697760 AGCCCGACAUUGUCAUUUAUUCAAUCUGCCAUUAUUGAGUUUGGUAAGUUGAGUGGGGUAUUCAGAAGUUUUG---UUCUUUGCCC---------CCCGAGUU (((.((............((((((..(((((..........)))))..))))))((((((...((((......---)))).)))))---------).)).))) ( -23.60, z-score = -2.28, R) >consensus AAACCAACAUUGCCAUUCACUCAAUGUACCAUUAUUGAGUUUGGUAAGUUGAGUGGGGUAUUCAGAAAAUUUGAAUUUCUUUGCUUA____CAGACCCAAAUU ....((((.((((((...((((((((......)))))))).))))))))))((..(((.((((((.....))))))..)))..)).................. (-20.51 = -21.40 + 0.89)
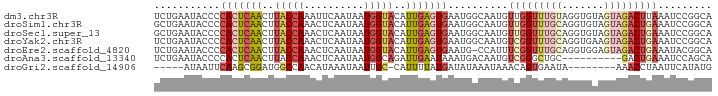
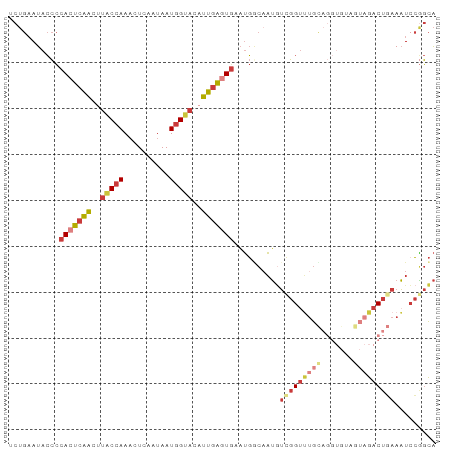
| Location | 22,398,181 – 22,398,274 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 75.58 |
| Shannon entropy | 0.49149 |
| G+C content | 0.39841 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -12.45 |
| Energy contribution | -14.17 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
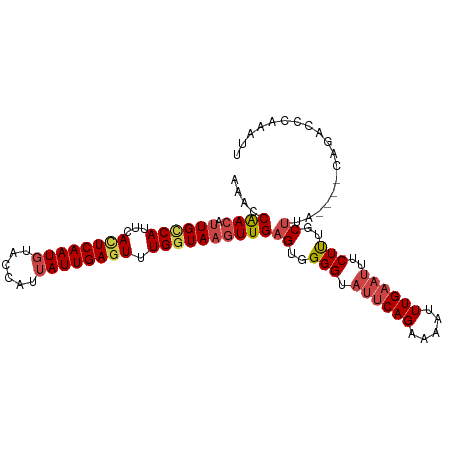
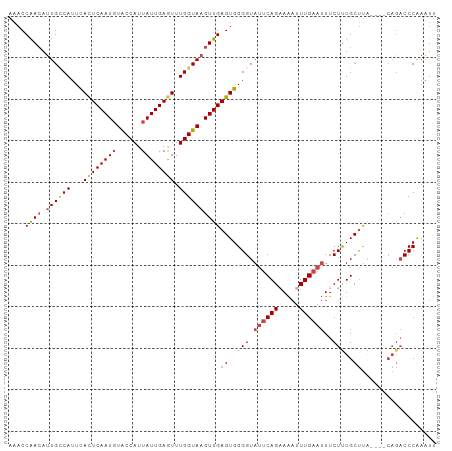
>dm3.chr3R 22398181 93 + 27905053 UCUGAAUACCCCACUCAACUUACCAAAUUCAAUAAUGGUACAUUGAGUGAAUGGCAAUGUUGGUUUGUAGGUGUAGUAGACUUAAAUCCGGCA ........((.(((((((..(((((..........)))))..)))))))...))...((((((....(((((.......)))))...)))))) ( -22.80, z-score = -1.07, R) >droSim1.chr3R 22226126 93 + 27517382 GCUGAAUACCCCACUCAACUUACCAAACUCAAUAAUGGUACAUUGAGUGAAUGGCAAUGUUGGUUUGCAGGUGUAGUAGACUGAAAUCCGGCA ((((....((.(((((((..(((((..........)))))..)))))))...)).....(..((((((.......))))))..)....)))). ( -26.40, z-score = -1.67, R) >droSec1.super_13 1173320 93 + 2104621 GCUGAAUACCCCACUCAACUUACCAAACUCAAUAAUGGUACAUUGAGUGAAUGGCAAUGUUGGUUUGCAGGUGUAGUAGACUGAAAUCCGGCA ((((....((.(((((((..(((((..........)))))..)))))))...)).....(..((((((.......))))))..)....)))). ( -26.40, z-score = -1.67, R) >droYak2.chr3R 4729462 93 - 28832112 UCUGAAUACCCCACUCAACUUACCAAACUCAAUAAUGGUACAUUGAGUGAAUGGCAAUGUCGGUUUGCAGGUGAAGUAGACUGAAAUCCGGCA ...........(((((((..(((((..........)))))..)))))))....((....(((((((((.......)))))))))......)). ( -25.90, z-score = -1.91, R) >droEre2.scaffold_4820 4812445 92 - 10470090 UCUGAAUACCCCACUCAACUUACCAAACUCAAUAAUGGUACAUUGAGUGAAUG-CCAUUUCGGUUUGCAGGUGGAGUAGACUGAAAUACGGCA ...........(((((((..(((((..........)))))..)))))))..((-((((((((((((((.......))))))))))))..)))) ( -31.00, z-score = -3.93, R) >droAna3.scaffold_13340 6753321 83 - 23697760 UCUGAAUACCCCACUCAACUUACCAAACUCAAUAAUGGCAGAUUGAAUAAAUGACAAUGUCGGGCUGC----------GACUGAAAUCCAGCA ..........((..((((.((.(((..........))).)).))))......(((...)))))((((.----------((......)))))). ( -11.60, z-score = 0.17, R) >droGri2.scaffold_14906 13008878 79 - 14172833 -----AUAAUUCAAGCGGAUGGCCAACAUAAAUAAUUGC-CAUUUUAUGAUAUAAAUAAACAGUGAAUA--------AAACCUAAUUCAUAUG -----.....(((...(((((((..............))-)))))..)))............((((((.--------.......))))))... ( -10.04, z-score = -0.61, R) >consensus UCUGAAUACCCCACUCAACUUACCAAACUCAAUAAUGGUACAUUGAGUGAAUGGCAAUGUCGGUUUGCAGGUGUAGUAGACUGAAAUCCGGCA ...........(((((((..(((((..........)))))..)))))))..........(((((((((.......)))))))))......... (-12.45 = -14.17 + 1.72)

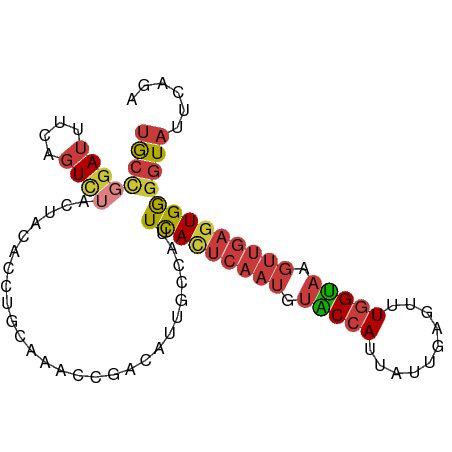
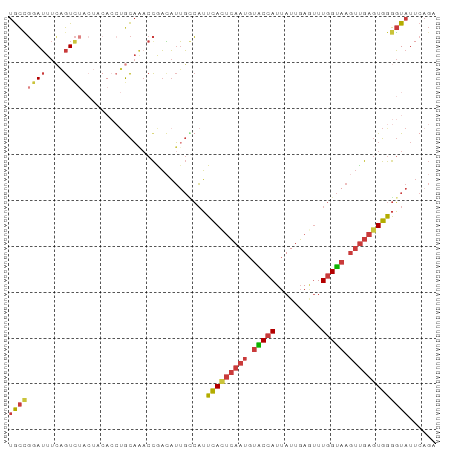
| Location | 22,398,181 – 22,398,274 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 75.58 |
| Shannon entropy | 0.49149 |
| G+C content | 0.39841 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -14.72 |
| Energy contribution | -15.87 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22398181 93 - 27905053 UGCCGGAUUUAAGUCUACUACACCUACAAACCAACAUUGCCAUUCACUCAAUGUACCAUUAUUGAAUUUGGUAAGUUGAGUGGGGUAUUCAGA ....((((....))))..(((.(((((....((((.((((((.....((((((......))))))...)))))))))).))))))))...... ( -22.40, z-score = -1.30, R) >droSim1.chr3R 22226126 93 - 27517382 UGCCGGAUUUCAGUCUACUACACCUGCAAACCAACAUUGCCAUUCACUCAAUGUACCAUUAUUGAGUUUGGUAAGUUGAGUGGGGUAUUCAGC ....((((....))))..(((.((..(....((((.((((((...((((((((......)))))))).)))))))))).)..)))))...... ( -26.40, z-score = -1.89, R) >droSec1.super_13 1173320 93 - 2104621 UGCCGGAUUUCAGUCUACUACACCUGCAAACCAACAUUGCCAUUCACUCAAUGUACCAUUAUUGAGUUUGGUAAGUUGAGUGGGGUAUUCAGC ....((((....))))..(((.((..(....((((.((((((...((((((((......)))))))).)))))))))).)..)))))...... ( -26.40, z-score = -1.89, R) >droYak2.chr3R 4729462 93 + 28832112 UGCCGGAUUUCAGUCUACUUCACCUGCAAACCGACAUUGCCAUUCACUCAAUGUACCAUUAUUGAGUUUGGUAAGUUGAGUGGGGUAUUCAGA ((.(((.((((((..........))).)))))).)).((((...((((((((.(((((..........))))).)))))))).))))...... ( -25.50, z-score = -1.47, R) >droEre2.scaffold_4820 4812445 92 + 10470090 UGCCGUAUUUCAGUCUACUCCACCUGCAAACCGAAAUGG-CAUUCACUCAAUGUACCAUUAUUGAGUUUGGUAAGUUGAGUGGGGUAUUCAGA ............(..(((((((((.((..((((((....-..)))((((((((......))))))))..)))..)).).))))))))..)... ( -24.90, z-score = -1.45, R) >droAna3.scaffold_13340 6753321 83 + 23697760 UGCUGGAUUUCAGUC----------GCAGCCCGACAUUGUCAUUUAUUCAAUCUGCCAUUAUUGAGUUUGGUAAGUUGAGUGGGGUAUUCAGA ..((((((.((.(((----------(.....))))........((((((((..(((((..........)))))..)))))))))).)))))). ( -23.00, z-score = -1.56, R) >droGri2.scaffold_14906 13008878 79 + 14172833 CAUAUGAAUUAGGUUU--------UAUUCACUGUUUAUUUAUAUCAUAAAAUG-GCAAUUAUUUAUGUUGGCCAUCCGCUUGAAUUAU----- ..(((((((..(((..--------.....)))....)))))))(((....(((-(((((.......))).))))).....))).....----- ( -9.90, z-score = 0.55, R) >consensus UGCCGGAUUUCAGUCUACUACACCUGCAAACCGACAUUGCCAUUCACUCAAUGUACCAUUAUUGAGUUUGGUAAGUUGAGUGGGGUAUUCAGA ((((((((....))))...........................(((((((((.(((((..........))))).)))))))))))))...... (-14.72 = -15.87 + 1.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:10 2011