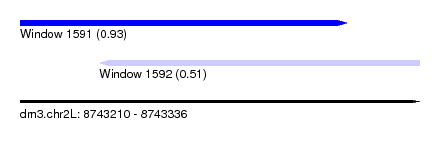
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,743,210 – 8,743,336 |
| Length | 126 |
| Max. P | 0.931454 |

| Location | 8,743,210 – 8,743,313 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.05 |
| Shannon entropy | 0.56739 |
| G+C content | 0.51060 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -17.35 |
| Energy contribution | -16.67 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
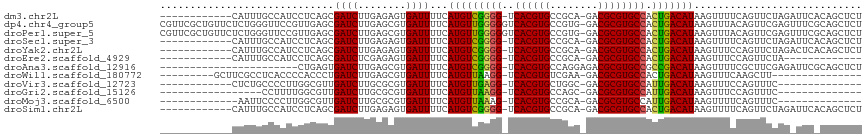
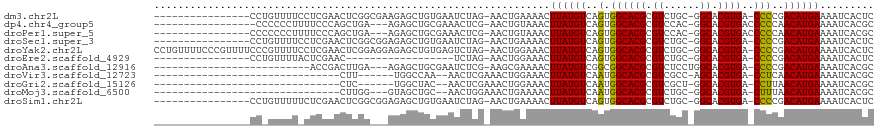
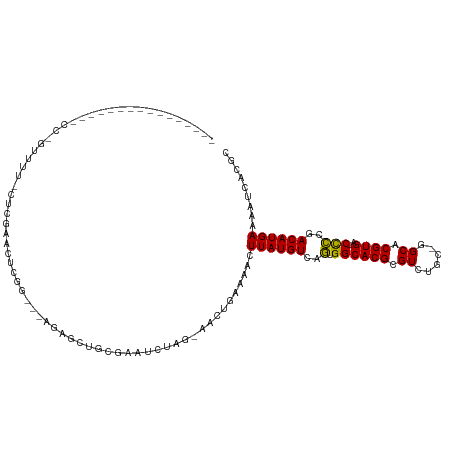
>dm3.chr2L 8743210 103 + 23011544 ------------CAUUUGCCAUCCUCAGCGAUCUUGAGAGUGAUUUUCAUGUCGGGG-UCACGUGCCGCA-GACGCGUGCCACUGACAUAAGUUUUCAGUUCUAGAUUCACAGCUCU ------------..............((((((((.(((..(((..((.(((((((((-.((((((.....-..)))))))).))))))).))...))).))).)))))....))).. ( -28.40, z-score = -0.68, R) >dp4.chr4_group5 1526473 116 - 2436548 CGUUCGCUGUUCUCUGGGUUCCGUUGAGCGAUCUUGAGCGUGAUUUUCAUGUUGGGGGUCACGUGCCGUG-GACGCGUGCCACUGACAUAAGUUUACAGUUCGAGUUUCGCAGCUCU .....(((..((..(((...)))..))((((.(((((((((((.(((.((((..(.((.(((((((....-).)))))))).)..))))))).)))).)))))))..)))))))... ( -41.60, z-score = -1.42, R) >droPer1.super_5 1494739 116 - 6813705 CGUUCGCUGUUCUCUGGGUUCCGUUGAGCGAUCUUGAGCGUGAUUUUCAUGUUGGGGGUCACGUGCCGUG-GACGCGUGCCACUGACAUAAGUUUACAGUUCGAGUUUCGCAGCUCU .....(((..((..(((...)))..))((((.(((((((((((.(((.((((..(.((.(((((((....-).)))))))).)..))))))).)))).)))))))..)))))))... ( -41.60, z-score = -1.42, R) >droSec1.super_3 4216556 103 + 7220098 ------------CAUUUGCCAUCCUCAGCGAUCUUGAGAGUGAUUUUCAUGUCGGGG-UCACGUGCCGCA-GACGCGUGCCACUGACAUAAGUUUUCAGUUCUAGAUUCACAGCUCU ------------..............((((((((.(((..(((..((.(((((((((-.((((((.....-..)))))))).))))))).))...))).))).)))))....))).. ( -28.40, z-score = -0.68, R) >droYak2.chr2L 11384914 103 + 22324452 ------------CAUUUGCCAUCCUCAGCGAUCUUGAGAGUGAUUUUCAUGUCGGGG-UCACGUGCCGCA-GACGCGUGCCACUGACAUAAGUUUCCAGUUCUAGACUCACAGCUCU ------------.....((....(((((.....))))).((((.....(((((((((-.((((((.....-..)))))))).)))))))..((((........)))))))).))... ( -28.30, z-score = -0.38, R) >droEre2.scaffold_4929 9329676 90 + 26641161 ------------CAUUUGCCAUCCUCAGCGAUCUCGAGAGUGAUUUUCAUGUCGGGG-UCACGUGCCGCA-GACGCGUGCCACUGACAUAAGUUUCCAGUUCUA------------- ------------...((((........))))....(((...((..((.(((((((((-.((((((.....-..)))))))).))))))).))..))...)))..------------- ( -24.90, z-score = -0.53, R) >droAna3.scaffold_12916 10381373 93 + 16180835 -----------------------CUGAGUGAUCUUGAGCGUGAUUUUCAUGUCGGGG-UCACGUGCCAGGAGACGCGUGCCGCCGACAUAAGUUUUCGCUUCGAGAUUCGCAGCUCU -----------------------..(((((((((((((((.(((((..(((((((((-.((((((........)))))))).))))))))))))..))).))))))))....)))). ( -40.10, z-score = -3.11, R) >droWil1.scaffold_180772 16818 91 + 8906247 ---------GCUUCGCCUCACCCCACCCUGAUCUUGAGCGUGAUUUUCAUGUUAAGG-UCACGUGUCGAA-GACGCGUGCCACUGACAUAAGUUUCAAGCUU--------------- ---------(((((((((((..............)))).)))).....((((((.((-.((((((((...-))))))))))..))))))........)))..--------------- ( -25.64, z-score = -2.05, R) >droVir3.scaffold_12723 4727958 89 + 5802038 ------------CUCUGCCCCUUGGCGUUGAUCUUGCGCGUGAUUUUCAUGUUGAGG-UCACGUGCUGGC-GACGCGUGCCAUUGACAUAAGUUUCCAGUUUC-------------- ------------....(((....)))((..((...((((((((((((......))))-)))))))).(((-(.....))))))..))................-------------- ( -27.60, z-score = -1.13, R) >droGri2.scaffold_15126 7731662 84 + 8399593 -----------------CCUUUUGGCGUUGAUCUUGCGCGUGAUUUUCAUGUUAAGG-UCACGUGCCAGC-GACGCGUGCCAUUGACAUAAGUUUCCAGUUUC-------------- -----------------.......((((((..((.((((((((((((......))))-)))))))).)))-)))))...........................-------------- ( -25.00, z-score = -1.73, R) >droMoj3.scaffold_6500 10695755 88 + 32352404 -------------AAUUCCCCUUGGCGUUGAUCUUGCGCGUGAUUUUCAUGUUAAAG-UCACGUGCCGCA-GACGCGUGCCAUUGACAUAAGUUUUCAGUUUC-------------- -------------......(..(((((........((((((((((((......))))-))))))))(((.-...))))))))..)..................-------------- ( -24.50, z-score = -1.99, R) >droSim1.chr2L 8522112 103 + 22036055 ------------CAUUUGCCAUCCUCAGCGAUCUUGAGAGUGAUUUUCAUGUCGGGG-UCACGUGCCGCA-GACGCGUGCCACUGACAUAAGUUUUCAGUUCUAGAUUCACAGCUCU ------------..............((((((((.(((..(((..((.(((((((((-.((((((.....-..)))))))).))))))).))...))).))).)))))....))).. ( -28.40, z-score = -0.68, R) >consensus ____________CAUUUGCCAUCCUCAGCGAUCUUGAGCGUGAUUUUCAUGUCGGGG_UCACGUGCCGCA_GACGCGUGCCACUGACAUAAGUUUCCAGUUCUAG_UUC_CAGCUCU .............................((((........))))...(((((((((..((((((........)))))))).)))))))............................ (-17.35 = -16.67 + -0.68)
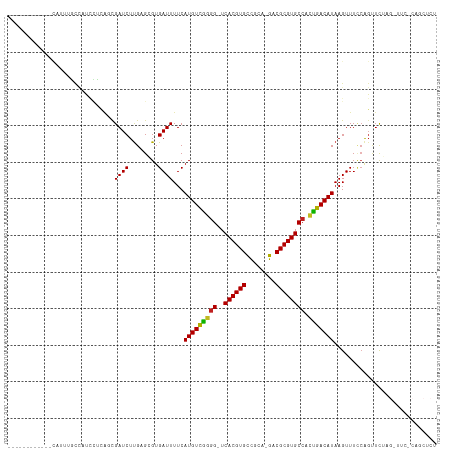
| Location | 8,743,235 – 8,743,336 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.54 |
| Shannon entropy | 0.52396 |
| G+C content | 0.52016 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -12.30 |
| Energy contribution | -11.91 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.510348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
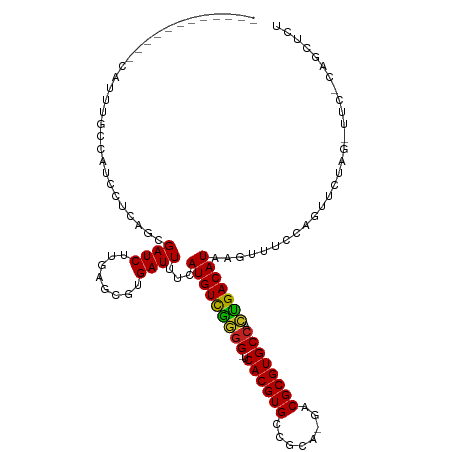
>dm3.chr2L 8743235 101 - 23011544 ----------------CCUGUUUUCCUCGAACUCGGCGAAGAGCUGUGAAUCUAG-AACUGAAAACUUAUGUCAGUGGCACGCGUCUGC-GGCACGUGA-CCCCGACAUGAAAAUCACUC ----------------...((((((((.((...((((.....))))....)).))-....))))))(((((((.(.((((((.((....-.)).)))).-))).)))))))......... ( -27.30, z-score = -0.98, R) >dp4.chr4_group5 1526510 98 + 2436548 -----------------CCCCCCUUUUCCCAGCUGA---AGAGCUGCGAAACUCG-AACUGUAAACUUAUGUCAGUGGCACGCGUCCAC-GGCACGUGACCCCCAACAUGAAAAUCACGC -----------------.......((((.(((((..---..))))).))))....-..........((((((..(.((((((.((....-.)).)))).)).)..))))))......... ( -23.40, z-score = -1.30, R) >droPer1.super_5 1494776 99 + 6813705 ----------------CCCCCCCUUUUCCCAGCUGA---AGAGCUGCGAAACUCG-AACUGUAAACUUAUGUCAGUGGCACGCGUCCAC-GGCACGUGACCCCCAACAUGAAAAUCACGC ----------------........((((.(((((..---..))))).))))....-..........((((((..(.((((((.((....-.)).)))).)).)..))))))......... ( -23.40, z-score = -1.43, R) >droSec1.super_3 4216581 101 - 7220098 ----------------CCUGUUUUCCUCGAACUCGGCGGAGAGCUGUGAAUCUAG-AACUGAAAACUUAUGUCAGUGGCACGCGUCUGC-GGCACGUGA-CCCCGACAUGAAAAUCACUC ----------------...((((((((((....))).))))))).((((.((...-....))....(((((((.(.((((((.((....-.)).)))).-))).)))))))...)))).. ( -31.80, z-score = -1.93, R) >droYak2.chr2L 11384939 117 - 22324452 CCUGUUUUCCCGUUUUCCCGUUUUCCUCGAACUCGGAGGAGAGCUGUGAGUCUAG-AACUGGAAACUUAUGUCAGUGGCACGCGUCUGC-GGCACGUGA-CCCCGACAUGAAAAUCACUC .....(((((.(((((((.(((((((((.......))))))))).).)).....)-))).))))).(((((((.(.((((((.((....-.)).)))).-))).)))))))......... ( -37.30, z-score = -1.42, R) >droEre2.scaffold_4929 9329701 83 - 26641161 ----------------CCUGUUUUACUCGAAC------------------UCUAG-AACUGGAAACUUAUGUCAGUGGCACGCGUCUGC-GGCACGUGA-CCCCGACAUGAAAAUCACUC ----------------((.((((((.......------------------..)))-))).))....(((((((.(.((((((.((....-.)).)))).-))).)))))))......... ( -22.30, z-score = -1.71, R) >droAna3.scaffold_12916 10381387 89 - 16180835 --------------------------ACCGACUUGA---AGAGCUGCGAAUCUCG-AAGCGAAAACUUAUGUCGGCGGCACGCGUCUCCUGGCACGUGA-CCCCGACAUGAAAAUCACGC --------------------------.......(((---...(((.((.....))-.)))......(((((((((.((((((.(((....))).)))).-)))))))))))...)))... ( -32.00, z-score = -2.92, R) >droVir3.scaffold_12723 4727983 79 - 5802038 -------------------------------CUU------UGGCCAA--AACUCGAAACUGGAAACUUAUGUCAAUGGCACGCGUCGCC-AGCACGUGA-CCUCAACAUGAAAAUCACGC -------------------------------..(------((((...--.....((....(....).....))...(((....))))))-))..(((((-..((.....))...))))). ( -15.00, z-score = 0.08, R) >droGri2.scaffold_15126 7731682 79 - 8399593 -------------------------------CUC------UGGCUAC--AACUCGAAACUGGAAACUUAUGUCAAUGGCACGCGUCGCU-GGCACGUGA-CCUUAACAUGAAAAUCACGC -------------------------------...------.......--.....((....(....)((((((.((.((((((.((....-.)).)))).-)))).))))))...)).... ( -15.60, z-score = -0.41, R) >droMoj3.scaffold_6500 10695779 82 - 32352404 -------------------------------CUUGG---GUAGCUGC--AACUGGAAACUGAAAACUUAUGUCAAUGGCACGCGUCUGC-GGCACGUGA-CUUUAACAUGAAAAUCACGC -------------------------------.....---...(((((--(((.(....)..........(((.....)))...)).)))-))).(((((-.(((......))).))))). ( -17.80, z-score = -0.17, R) >droSim1.chr2L 8522137 101 - 22036055 ----------------CCUGUUUUUCUCGAACUCGGCGGAGAGCUGUGAAUCUAG-AACUGAAAACUUAUGUCAGUGGCACGCGUCUGC-GGCACGUGA-CCCCGACAUGAAAAUCACUC ----------------...((((((((((....))).))))))).((((.((...-....))....(((((((.(.((((((.((....-.)).)))).-))).)))))))...)))).. ( -28.90, z-score = -1.01, R) >consensus ________________CC_GUUUU_CUCGAACUCGG___AGAGCUGCGAAUCUAG_AACUGAAAACUUAUGUCAGUGGCACGCGUCUGC_GGCACGUGA_CCCCGACAUGAAAAUCACGC ..................................................................((((((..(.((((((.(((....))).))))..)))..))))))......... (-12.30 = -11.91 + -0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:49 2011