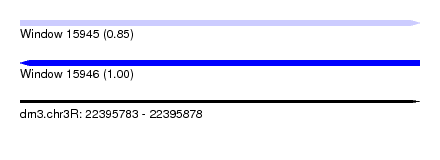
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,395,783 – 22,395,878 |
| Length | 95 |
| Max. P | 0.995199 |

| Location | 22,395,783 – 22,395,878 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 92.76 |
| Shannon entropy | 0.11846 |
| G+C content | 0.33009 |
| Mean single sequence MFE | -15.00 |
| Consensus MFE | -12.39 |
| Energy contribution | -12.95 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
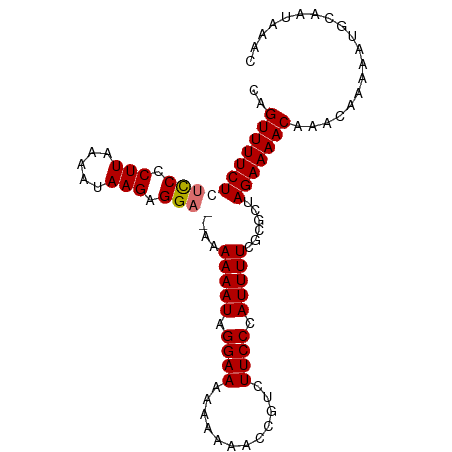
>dm3.chr3R 22395783 95 + 27905053 CAGUUUUCUCUCCCCUUAAAAUAAGAGGA--AAAAAAUAGGAAAAAAAAACCAUCUUCCCAUUUUCGCGCUAGAAAACAAACAAAAAUGCAAUAAAC ..(((((((.(((.((((...)))).)))--..(((((.((((............)))).)))))......)))))))................... ( -13.40, z-score = -2.30, R) >droSim1.chr3R 22223728 97 + 27517382 CAGUUUUCUCUUCCCUUAAAAUAAGAGGAGAAAAAAAUAGGAAAAAAAAACGGUCUUCCCAUUUUCGCGCUAGAAAACAAACAAAAAUGCAAUAAAC ..(((((((((((.((((...)))).)))))..(((((.((((............)))).))))).......))))))................... ( -14.50, z-score = -1.27, R) >droSec1.super_13 1170943 96 + 2104621 CAGUUUUCUCUCCCCUUAAAAUAAGAGGAGAAAAAAAUAGGAAAAAAAAGC-GUCUUCCCAUUUUCGCGCUAGAAAACAAACAAAAAUGCAAUAAAC ..(((((((((((.((((...)))).))))).................(((-((............))))).))))))................... ( -18.70, z-score = -3.12, R) >droEre2.scaffold_4820 4810070 91 - 10470090 CAGUUUUCUCCCCCCUUUAAAUAAGAGGA---CAAAAUAGGAAAAAAA---CGUCUUCCCAUUUUCGCGCUAGAAAACAAACAAAAAUGCAAUAAAC ..(((((((....(((((.....))))).---.(((((.((((.....---....)))).)))))......)))))))................... ( -13.40, z-score = -1.91, R) >consensus CAGUUUUCUCUCCCCUUAAAAUAAGAGGA__AAAAAAUAGGAAAAAAAAACCGUCUUCCCAUUUUCGCGCUAGAAAACAAACAAAAAUGCAAUAAAC ..(((((((((((.(((.....))).))))...(((((.((((............)))).)))))......)))))))................... (-12.39 = -12.95 + 0.56)

| Location | 22,395,783 – 22,395,878 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 92.76 |
| Shannon entropy | 0.11846 |
| G+C content | 0.33009 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -23.74 |
| Energy contribution | -24.55 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.995199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
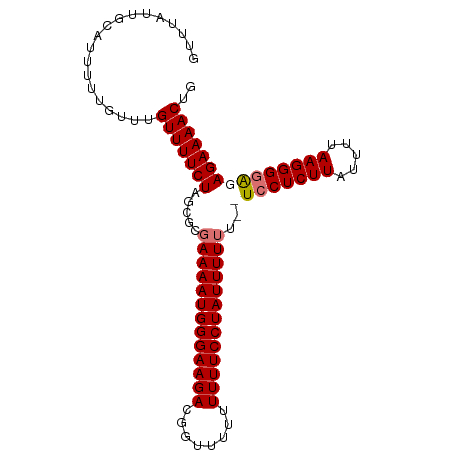
>dm3.chr3R 22395783 95 - 27905053 GUUUAUUGCAUUUUUGUUUGUUUUCUAGCGCGAAAAUGGGAAGAUGGUUUUUUUUUCCUAUUUUUU--UCCUCUUAUUUUAAGGGGAGAGAAAACUG ...................(((((((.....(((((((((((((........)))))))))))))(--((((((((...)))))))))))))))).. ( -27.20, z-score = -3.65, R) >droSim1.chr3R 22223728 97 - 27517382 GUUUAUUGCAUUUUUGUUUGUUUUCUAGCGCGAAAAUGGGAAGACCGUUUUUUUUUCCUAUUUUUUUCUCCUCUUAUUUUAAGGGAAGAGAAAACUG ((((.((.(((((((((..(((....)))))))))))).)))))).(((((((((((((.......................))))))))))))).. ( -22.40, z-score = -1.99, R) >droSec1.super_13 1170943 96 - 2104621 GUUUAUUGCAUUUUUGUUUGUUUUCUAGCGCGAAAAUGGGAAGAC-GCUUUUUUUUCCUAUUUUUUUCUCCUCUUAUUUUAAGGGGAGAGAAAACUG ...................((((((......(((((((((((((.-......))))))))))))).((((((((((...)))))))))))))))).. ( -28.60, z-score = -3.95, R) >droEre2.scaffold_4820 4810070 91 + 10470090 GUUUAUUGCAUUUUUGUUUGUUUUCUAGCGCGAAAAUGGGAAGACG---UUUUUUUCCUAUUUUG---UCCUCUUAUUUAAAGGGGGGAGAAAACUG ((((.((.(((((((((..(((....)))))))))))).))))))(---((((((((((.(((((---..........))))).))))))))))).. ( -24.10, z-score = -2.50, R) >consensus GUUUAUUGCAUUUUUGUUUGUUUUCUAGCGCGAAAAUGGGAAGACGGUUUUUUUUUCCUAUUUUUU__UCCUCUUAUUUUAAGGGGAGAGAAAACUG ...................(((((((.....(((((((((((((........)))))))))))))..((((((((.....))))))))))))))).. (-23.74 = -24.55 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:06 2011