| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,382,080 – 22,382,178 |
| Length | 98 |
| Max. P | 0.989084 |

| Location | 22,382,080 – 22,382,178 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 70.03 |
| Shannon entropy | 0.57720 |
| G+C content | 0.26885 |
| Mean single sequence MFE | -19.13 |
| Consensus MFE | -11.13 |
| Energy contribution | -11.34 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.989084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
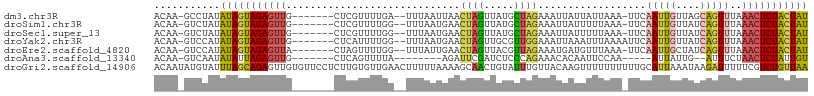
>dm3.chr3R 22382080 98 + 27905053 ACAA-GCCUAUAUAGUAGAGUUG-------CUCGUUUUGA--UUUAAUUAACUAGUUAUGCUAGAAAUUAUUAUUAAA-UUCAAUUGUUAGCAGUUUAAACUCUACUAU ....-......(((((((((((.-------.....(((((--(.(((((..((((.....)))).)))))..))))))-...(((((....)))))..))))))))))) ( -20.70, z-score = -2.08, R) >droSim1.chr3R 22216589 98 + 27517382 ACAA-GUCUAUAUAGUAGAGUUG-------CUCGUUUUGG--UUUAAUGAACUAGUUAUGCUAGAAAUUAUUUUUAAA-UUCAAUUGUUAUCAGUUUAAACUCUACUAU ....-......(((((((((((.-------....((((((--(.((((......)))).)))))))............-...(((((....)))))..))))))))))) ( -20.30, z-score = -2.13, R) >droSec1.super_13 1164496 98 + 2104621 ACAA-GUCUAUAUAGUAGAGUUG-------CUCGUUUUGG--UUUAAUGAACUAGUUAUGCUAGAAAUUAUUUUUAAA-UUCAAUUGUUAUCAGUUUAAACUCUACUAU ....-......(((((((((((.-------....((((((--(.((((......)))).)))))))............-...(((((....)))))..))))))))))) ( -20.30, z-score = -2.13, R) >droYak2.chr3R 4720364 99 - 28832112 ACAA-GUCCAUAUAGUAGAGUUG-------CUCAUUUUGG--UUUAAUGAACUAGUUGCGUUGGAAAUUAAAUUUAAAAUUCAAUUGUUAUCAGUUUAAACUCUACUAU ....-......((((((((((((-------((......))--)....((((((.((.((((((((..(((....)))..))))).))).)).))))))))))))))))) ( -19.80, z-score = -2.01, R) >droEre2.scaffold_4820 4803581 98 - 10470090 ACAA-GUCCAUAUAGUAGAGUUA-------CUAGUUUUGG--UUUAUUGAACUAGUUACGUUAGAAAUGAUGUUUAAA-UUCAAUUGCUAUCAGUUUAAACUCUACUAU ....-......((((((((((((-------((.(...(((--(..((((((......((((((....)))))).....-)))))).)))).))))...))))))))))) ( -25.10, z-score = -3.40, R) >droAna3.scaffold_13340 6744543 86 - 23697760 ACAA-GUCAAUAUAUUAGAGUUG-------CUCAGUUUUA--------AGAUUCGAUCUCCCAGAAACACAAUUCCAA-----AUUAUUG--AUUUCUAACUCUAUUGU ..((-(((((((.....((((((-------....(((((.--------((((...))))....))))).))))))...-----..)))))--))))............. ( -12.20, z-score = -1.09, R) >droGri2.scaffold_14906 12998092 109 - 14172833 ACAAUAUGUAUUUAGCAGAGUUGUGUUCCUCUUGUGUUGAACUUUUUAAAAGCAACUGUAUUUGUUACAAGUUUUUUUUUUGCAUUAAAUAAGAUUUUUCGUCUGUUAA ...........(((((((((((((((((..(....)..)))).........)))))..((((((...((((.......))))...))))))..........)))))))) ( -15.50, z-score = 0.36, R) >consensus ACAA_GUCUAUAUAGUAGAGUUG_______CUCGUUUUGG__UUUAAUGAACUAGUUAUGCUAGAAAUUAUUUUUAAA_UUCAAUUGUUAUCAGUUUAAACUCUACUAU ...........(((((((((((.............................((((.....))))..................(((((....)))))..))))))))))) (-11.13 = -11.34 + 0.21)
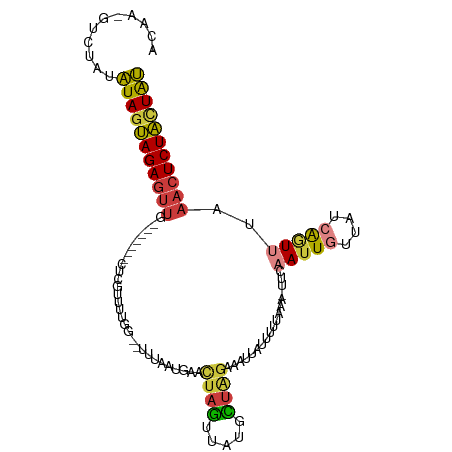
| Location | 22,382,080 – 22,382,178 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 70.03 |
| Shannon entropy | 0.57720 |
| G+C content | 0.26885 |
| Mean single sequence MFE | -16.52 |
| Consensus MFE | -6.84 |
| Energy contribution | -7.21 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22382080 98 - 27905053 AUAGUAGAGUUUAAACUGCUAACAAUUGAA-UUUAAUAAUAAUUUCUAGCAUAACUAGUUAAUUAAA--UCAAAACGAG-------CAACUCUACUAUAUAGGC-UUGU ((((((((((......((((.....((((.-((((((........((((.....))))...))))))--))))....))-------))))))))))))......-.... ( -19.70, z-score = -2.39, R) >droSim1.chr3R 22216589 98 - 27517382 AUAGUAGAGUUUAAACUGAUAACAAUUGAA-UUUAAAAAUAAUUUCUAGCAUAACUAGUUCAUUAAA--CCAAAACGAG-------CAACUCUACUAUAUAGAC-UUGU (((((((((((................(((-.(((....))).)))...........((((......--.......)))-------))))))))))))......-.... ( -15.52, z-score = -1.77, R) >droSec1.super_13 1164496 98 - 2104621 AUAGUAGAGUUUAAACUGAUAACAAUUGAA-UUUAAAAAUAAUUUCUAGCAUAACUAGUUCAUUAAA--CCAAAACGAG-------CAACUCUACUAUAUAGAC-UUGU (((((((((((................(((-.(((....))).)))...........((((......--.......)))-------))))))))))))......-.... ( -15.52, z-score = -1.77, R) >droYak2.chr3R 4720364 99 + 28832112 AUAGUAGAGUUUAAACUGAUAACAAUUGAAUUUUAAAUUUAAUUUCCAACGCAACUAGUUCAUUAAA--CCAAAAUGAG-------CAACUCUACUAUAUGGAC-UUGU (((((((((((............((((((((.....)))))))).............(((((((...--....))))))-------))))))))))))......-.... ( -21.10, z-score = -3.58, R) >droEre2.scaffold_4820 4803581 98 + 10470090 AUAGUAGAGUUUAAACUGAUAGCAAUUGAA-UUUAAACAUCAUUUCUAACGUAACUAGUUCAAUAAA--CCAAAACUAG-------UAACUCUACUAUAUGGAC-UUGU (((((((((((........(((.((.(((.-........))).))))).....(((((((.......--....))))))-------))))))))))))......-.... ( -18.70, z-score = -2.17, R) >droAna3.scaffold_13340 6744543 86 + 23697760 ACAAUAGAGUUAGAAAU--CAAUAAU-----UUGGAAUUGUGUUUCUGGGAGAUCGAAUCU--------UAAAACUGAG-------CAACUCUAAUAUAUUGAC-UUGU ................(--(((((..-----(((((.((((((((.(((((.......)))--------))))))...)-------))).)))))..)))))).-.... ( -14.20, z-score = -0.22, R) >droGri2.scaffold_14906 12998092 109 + 14172833 UUAACAGACGAAAAAUCUUAUUUAAUGCAAAAAAAAAACUUGUAACAAAUACAGUUGCUUUUAAAAAGUUCAACACAAGAGGAACACAACUCUGCUAAAUACAUAUUGU (((.((((............(((((.((((.........(((((.....)))))))))..)))))..((((..(....)..)))).....)))).)))........... ( -10.90, z-score = 0.25, R) >consensus AUAGUAGAGUUUAAACUGAUAACAAUUGAA_UUUAAAAAUAAUUUCUAGCAUAACUAGUUCAUUAAA__CCAAAACGAG_______CAACUCUACUAUAUAGAC_UUGU (((((((((((............................................................................)))))))))))........... ( -6.84 = -7.21 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:46:03 2011