| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,367,490 – 22,367,584 |
| Length | 94 |
| Max. P | 0.939503 |

| Location | 22,367,490 – 22,367,584 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Shannon entropy | 0.14813 |
| G+C content | 0.43817 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -27.44 |
| Energy contribution | -28.28 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
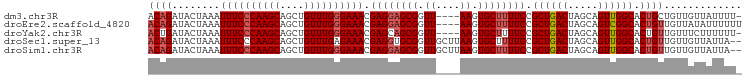
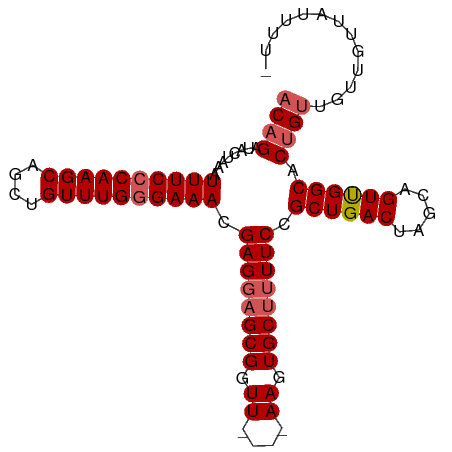
>dm3.chr3R 22367490 94 + 27905053 ACAGAUACUAAAUUUCCCAAGCAGCUGUUUGGGAAACGAGGAGCGGUU----AAGUGCUUUUCCGCUGACUAGCAGUUGGCACUGCUGUUGUUAUUUU- .(((........((((((((((....)))))))))).((((((((...----...))))))))..)))..(((((((....)))))))..........- ( -31.20, z-score = -2.18, R) >droEre2.scaffold_4820 4786570 95 - 10470090 ACAGAUACUAAAUUUCCCAAGCAGCUGUUUGGGAAACGAGGAGCGGUU----AAGUGCUUUUCCGCUGACUAGCAGUCGGCACUGUUGUUAUAUUUUUU ((((........((((((((((....)))))))))).((((((((...----...)))))))).((((((.....)))))).))))............. ( -33.30, z-score = -3.52, R) >droYak2.chr3R 4705276 94 - 28832112 ACUGAUACUAAAUUUCCCAAGCAGCUGUUUGGGAAACGAGCAGCGGUU----AAGUGCUUUUCCGCUGACUAGCAGUUGGCACUGUUGUUUCUUUUUU- ............((((((((((....)))))))))).(((((((((((----((.((((..((....))..)))).)))..)))))))))).......- ( -30.70, z-score = -2.37, R) >droSec1.super_13 1150071 97 + 2104621 ACAGAUACUAAAUUUCCCAAGCAGCUGUUUGAGAAACGAGGUGCGGUUGCUUAAGUGCUUUUCCGCUGACUAGCAGUUGGCACUGUUGUUGUUAUUA-- ((((..((..........(((((((((((((.....)))...)))))))))).((((((...(.(((....))).)..))))))))..)))).....-- ( -23.40, z-score = 0.40, R) >droSim1.chr3R 22202061 97 + 27517382 ACAGAUACUAAAUUUCCCAAGCAGCUGUUUGGGAAACGAGGAGCGGUUGCUUAAGUGCUUUUCCGCUGACUAGCAGUUGGCACUGUUGUUGUUAUUA-- ((((........((((((((((....)))))))))).((((((((.((....)).)))))))).((..((.....))..)).))))...........-- ( -31.90, z-score = -2.07, R) >consensus ACAGAUACUAAAUUUCCCAAGCAGCUGUUUGGGAAACGAGGAGCGGUU____AAGUGCUUUUCCGCUGACUAGCAGUUGGCACUGUUGUUGUUAUUUU_ ((((........((((((((((....)))))))))).((((((((.((....)).)))))))).((((((.....)))))).))))............. (-27.44 = -28.28 + 0.84)
| Location | 22,367,490 – 22,367,584 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 91.25 |
| Shannon entropy | 0.14813 |
| G+C content | 0.43817 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -20.68 |
| Energy contribution | -20.36 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22367490 94 - 27905053 -AAAAUAACAACAGCAGUGCCAACUGCUAGUCAGCGGAAAAGCACUU----AACCGCUCCUCGUUUCCCAAACAGCUGCUUGGGAAAUUUAGUAUCUGU -...........((((((....))))))....(((((..........----..)))))....(((((((((.(....).)))))))))........... ( -23.90, z-score = -1.56, R) >droEre2.scaffold_4820 4786570 95 + 10470090 AAAAAAUAUAACAACAGUGCCGACUGCUAGUCAGCGGAAAAGCACUU----AACCGCUCCUCGUUUCCCAAACAGCUGCUUGGGAAAUUUAGUAUCUGU .............(((((((.(((.....)))(((((..........----..)))))....(((((((((.(....).)))))))))...))).)))) ( -22.50, z-score = -1.52, R) >droYak2.chr3R 4705276 94 + 28832112 -AAAAAAGAAACAACAGUGCCAACUGCUAGUCAGCGGAAAAGCACUU----AACCGCUGCUCGUUUCCCAAACAGCUGCUUGGGAAAUUUAGUAUCAGU -......((.((..((((....))))..((.((((((..........----..)))))))).(((((((((.(....).)))))))))...)).))... ( -24.20, z-score = -1.46, R) >droSec1.super_13 1150071 97 - 2104621 --UAAUAACAACAACAGUGCCAACUGCUAGUCAGCGGAAAAGCACUUAAGCAACCGCACCUCGUUUCUCAAACAGCUGCUUGGGAAAUUUAGUAUCUGU --.....(((...((.((((...(((((....)))))....((......))....))))...(((((((((.(....).)))))))))...))...))) ( -19.30, z-score = 0.04, R) >droSim1.chr3R 22202061 97 - 27517382 --UAAUAACAACAACAGUGCCAACUGCUAGUCAGCGGAAAAGCACUUAAGCAACCGCUCCUCGUUUCCCAAACAGCUGCUUGGGAAAUUUAGUAUCUGU --...........(((((((............(((((....((......))..)))))....(((((((((.(....).)))))))))...))).)))) ( -22.30, z-score = -1.01, R) >consensus _AAAAUAACAACAACAGUGCCAACUGCUAGUCAGCGGAAAAGCACUU____AACCGCUCCUCGUUUCCCAAACAGCUGCUUGGGAAAUUUAGUAUCUGU .............(((((((...(((((....)))))....)))))................(((((((((.(....).)))))))))...))...... (-20.68 = -20.36 + -0.32)

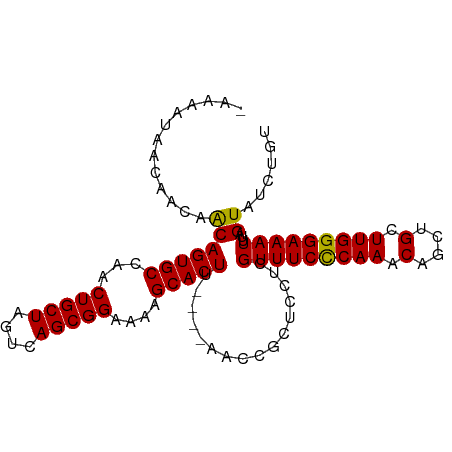
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:58 2011