
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,733,900 – 8,733,992 |
| Length | 92 |
| Max. P | 0.994357 |

| Location | 8,733,900 – 8,733,992 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.51 |
| Shannon entropy | 0.52855 |
| G+C content | 0.36751 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -14.13 |
| Energy contribution | -14.23 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.994357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
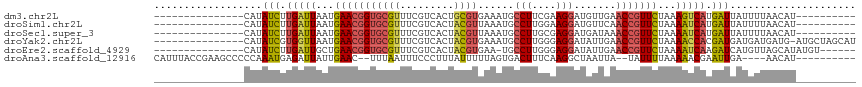
>dm3.chr2L 8733900 92 + 23011544 ---------------CAUAUCUUGAUUAAUGAACGGUGCGUUUCGUCACUGCGUGAAAUGCCUUCGAAGGAUGUUGAACCGUUCUAAAGUCAUGAUUAUUUUAACAU---------- ---------------...(((.(((((...(((((((((((((((........))))))))..((....))......)))))))...))))).)))...........---------- ( -24.60, z-score = -2.34, R) >droSim1.chr2L 8510829 92 + 22036055 ---------------CAUAUCUUGAUUAAUGAACGGUGCGUUUCGUCACUACGUUAAAUGCCUUGGAAGGAUGUUCAACCGUUCUAAAAUCAUGAUUAUUUUAACAU---------- ---------------...(((.(((((...((((((((((((((((....)))..))))))..((((......)))))))))))...))))).)))...........---------- ( -20.80, z-score = -1.94, R) >droSec1.super_3 4207308 92 + 7220098 ---------------CAUAUCUUGAUUAAUGAACGGUGCGUUUCGUCACUACGUUAAAUGCCUUGCGAGGAUGAUAAACCGUUCUAAAAUCAUGAUUAUUUUAACAU---------- ---------------...(((.(((((...(((((((.....(((((.((.(((..........))))))))))...)))))))...))))).)))...........---------- ( -24.20, z-score = -3.27, R) >droYak2.chr2L 11375360 101 + 22324452 ---------------CAUAUCGUGGUUAAUGAACGGUGCGUUUCGUCACUACGUGAAAUGCCUUGGGAGGAUAUUGAACCGUUCUAAAACCACGAUGAUGAUGAUG-AUGCUAGCAU ---------------((((((((((((...(((((((((((((((........))))))))((....))........)))))))...))))))))).)))......-.......... ( -31.70, z-score = -2.85, R) >droEre2.scaffold_4929 9319667 95 + 26641161 ---------------CAUAUCUUGAUUGCUGAACGGUGCGUUUCGUCACUACGUGAA-UGCCUUGGGAGGAUAUUGAACCGUUCUAAAAUCAAGAUCAUGUUAGCAUAUGU------ ---------------((((((((((((...(((((((.((..((.((.(((.((...-.))..))))).))...)).)))))))...))))))))).)))...........------ ( -27.80, z-score = -2.35, R) >droAna3.scaffold_12916 10373283 99 + 16180835 CAUUUACCGAAGCCCCCAAAUGAGAUUAUUGAAC--UUUAAUUUCCCUUUAUUUUUAGUGACUUUCAAGGCUAAUUA--UAUUUUAAAAACGAAUUGA----AACAU---------- ..........((((......(((((((((((((.--..(((.......)))..)))))))).))))).)))).....--...................----.....---------- ( -9.50, z-score = -0.08, R) >consensus _______________CAUAUCUUGAUUAAUGAACGGUGCGUUUCGUCACUACGUGAAAUGCCUUGGAAGGAUAUUGAACCGUUCUAAAAUCAUGAUUAUUUUAACAU__________ ..................(((.(((((...(((((((((((.........))))......(((....))).......)))))))...))))).)))..................... (-14.13 = -14.23 + 0.10)

| Location | 8,733,900 – 8,733,992 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 68.51 |
| Shannon entropy | 0.52855 |
| G+C content | 0.36751 |
| Mean single sequence MFE | -18.98 |
| Consensus MFE | -12.25 |
| Energy contribution | -11.88 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.972039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
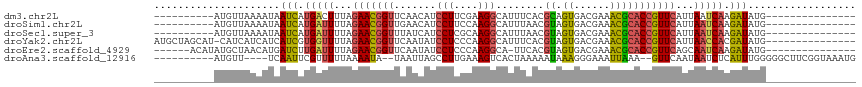
>dm3.chr2L 8733900 92 - 23011544 ----------AUGUUAAAAUAAUCAUGACUUUAGAACGGUUCAACAUCCUUCGAAGGCAUUUCACGCAGUGACGAAACGCACCGUUCAUUAAUCAAGAUAUG--------------- ----------...........(((.(((.....(((((((.(.....(((....)))....((((...))))......).))))))).....))).)))...--------------- ( -15.70, z-score = -0.46, R) >droSim1.chr2L 8510829 92 - 22036055 ----------AUGUUAAAAUAAUCAUGAUUUUAGAACGGUUGAACAUCCUUCCAAGGCAUUUAACGUAGUGACGAAACGCACCGUUCAUUAAUCAAGAUAUG--------------- ----------...........(((.(((((...(((((((((((...(((....)))..))))))...(((.((...))))))))))...))))).)))...--------------- ( -19.00, z-score = -1.80, R) >droSec1.super_3 4207308 92 - 7220098 ----------AUGUUAAAAUAAUCAUGAUUUUAGAACGGUUUAUCAUCCUCGCAAGGCAUUUAACGUAGUGACGAAACGCACCGUUCAUUAAUCAAGAUAUG--------------- ----------...........(((.(((((...(((((((......((.((((...((.......)).)))).)).....)))))))...))))).)))...--------------- ( -19.70, z-score = -2.04, R) >droYak2.chr2L 11375360 101 - 22324452 AUGCUAGCAU-CAUCAUCAUCAUCGUGGUUUUAGAACGGUUCAAUAUCCUCCCAAGGCAUUUCACGUAGUGACGAAACGCACCGUUCAUUAACCACGAUAUG--------------- ..........-......(((.(((((((((...(((((((.(.....(((....)))....((((...))))......).)))))))...))))))))))))--------------- ( -26.80, z-score = -3.38, R) >droEre2.scaffold_4929 9319667 95 - 26641161 ------ACAUAUGCUAACAUGAUCUUGAUUUUAGAACGGUUCAAUAUCCUCCCAAGGCA-UUCACGUAGUGACGAAACGCACCGUUCAGCAAUCAAGAUAUG--------------- ------...........((((.((((((((...(((((((.(.....(((....)))..-.((((...))))......).)))))))...))))))))))))--------------- ( -22.90, z-score = -2.54, R) >droAna3.scaffold_12916 10373283 99 - 16180835 ----------AUGUU----UCAAUUCGUUUUUAAAAUA--UAAUUAGCCUUGAAAGUCACUAAAAAUAAAGGGAAAUUAAA--GUUCAAUAAUCUCAUUUGGGGGCUUCGGUAAAUG ----------.....----.................((--(....((((((.(((....((........)).((.((((..--......)))).)).))).))))))...))).... ( -9.80, z-score = 1.07, R) >consensus __________AUGUUAAAAUAAUCAUGAUUUUAGAACGGUUCAACAUCCUCCCAAGGCAUUUAACGUAGUGACGAAACGCACCGUUCAUUAAUCAAGAUAUG_______________ .....................(((.(((((...(((((((.......(((....)))........((.((......)))))))))))...))))).))).................. (-12.25 = -11.88 + -0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:47 2011