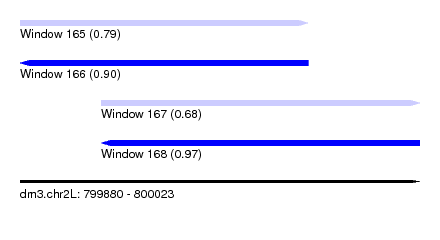
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 799,880 – 800,023 |
| Length | 143 |
| Max. P | 0.965697 |

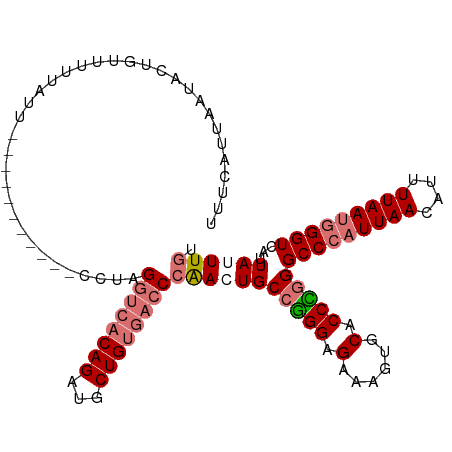
| Location | 799,880 – 799,983 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.80 |
| Shannon entropy | 0.29567 |
| G+C content | 0.42433 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -21.90 |
| Energy contribution | -23.77 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 799880 103 + 23011544 UUUCAUUAAUACUGUUUUUAUU-----------CCUAGGUAACAGAUGCUGUGACCCAACUGCCGGGAGAAAGUGCACCUGGGCCCAUUAACAUUUUAAUGGGUUAUUAUUUGU ......(((((...........-----------(((((((.((((...))))..(((.......))).........)))))))((((((((....)))))))).)))))..... ( -24.20, z-score = -0.41, R) >droYak2.chr2L 782627 108 + 22324452 UUUUAUUGAUCCUGUUUUUAAUAGCUCCUUGCACCUUGGUCACAGAUGCUGGG--CGGAGUGCAAGGUGGU----CACCUGGGCCGCUUAAGAUUUUAAUGGGUUAUUAUUUGU ..................((((((((((((((((.((.(((.(((...)))))--).)))))))))(((((----(.....)))))).............)))))))))..... ( -33.00, z-score = -1.44, R) >droSec1.super_14 769811 103 + 2068291 UUUCAUUAAUACUGUUUCUAUU-----------CCUAGGUCACAGAUGCUGUGACCCAACUGCCGGGCGAAAGUGCACCCGGGCCCAUUAACAUUUUAAUGGGUCAUUAUUUGU ......................-----------....((((((((...))))))))......(((((.(......).)))))(((((((((....))))))))).......... ( -31.80, z-score = -2.53, R) >droSim1.chr2L 801285 103 + 22036055 UUUCAUUAAUACUGUUCUUAUU-----------CCUAGGUCACAGAUGCUGUGACCCAAGUGCCGGGAGAAAGUGCACCCGGGCCCAUUAACGUUUUAAUGGGUCAUUAUUUGU ......................-----------....((((((((...))))))))((((((.((((.(......).))))((((((((((....))))))))))..)))))). ( -33.50, z-score = -2.80, R) >consensus UUUCAUUAAUACUGUUUUUAUU___________CCUAGGUCACAGAUGCUGUGACCCAACUGCCGGGAGAAAGUGCACCCGGGCCCAUUAACAUUUUAAUGGGUCAUUAUUUGU .....................................((((((((...))))))))(((((((((((.(......).)))))(((((((((....)))))))))...)))))). (-21.90 = -23.77 + 1.88)

| Location | 799,880 – 799,983 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.80 |
| Shannon entropy | 0.29567 |
| G+C content | 0.42433 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -21.11 |
| Energy contribution | -24.30 |
| Covariance contribution | 3.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 799880 103 - 23011544 ACAAAUAAUAACCCAUUAAAAUGUUAAUGGGCCCAGGUGCACUUUCUCCCGGCAGUUGGGUCACAGCAUCUGUUACCUAGG-----------AAUAAAAACAGUAUUAAUGAAA .............((((((.(((((....(((((((.(((.(........)))).)))))))..)))))(((((.......-----------......)))))..))))))... ( -23.52, z-score = -0.66, R) >droYak2.chr2L 782627 108 - 22324452 ACAAAUAAUAACCCAUUAAAAUCUUAAGCGGCCCAGGUG----ACCACCUUGCACUCCG--CCCAGCAUCUGUGACCAAGGUGCAAGGAGCUAUUAAAAACAGGAUCAAUAAAA .....(((((.(((.............((((.(.((((.----...)))).)....)))--)...((((((.......))))))..)).).))))).................. ( -21.00, z-score = -0.21, R) >droSec1.super_14 769811 103 - 2068291 ACAAAUAAUGACCCAUUAAAAUGUUAAUGGGCCCGGGUGCACUUUCGCCCGGCAGUUGGGUCACAGCAUCUGUGACCUAGG-----------AAUAGAAACAGUAUUAAUGAAA ...((((.((.((((((((....)))))))).(((((((......)))))))...(((((((((((...))))))))))).-----------........)).))))....... ( -36.20, z-score = -3.53, R) >droSim1.chr2L 801285 103 - 22036055 ACAAAUAAUGACCCAUUAAAACGUUAAUGGGCCCGGGUGCACUUUCUCCCGGCACUUGGGUCACAGCAUCUGUGACCUAGG-----------AAUAAGAACAGUAUUAAUGAAA ...((((.((.((((((((....)))))))).(((((.(......).)))))..((((((((((((...))))))))))))-----------........)).))))....... ( -35.80, z-score = -3.82, R) >consensus ACAAAUAAUAACCCAUUAAAAUGUUAAUGGGCCCAGGUGCACUUUCUCCCGGCACUUGGGUCACAGCAUCUGUGACCUAGG___________AAUAAAAACAGUAUUAAUGAAA ...........((((((((....)))))))).(((((((......)))))))..((((((((((((...))))))))))))................................. (-21.11 = -24.30 + 3.19)

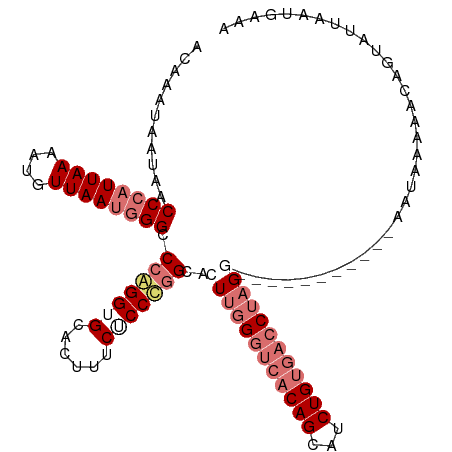
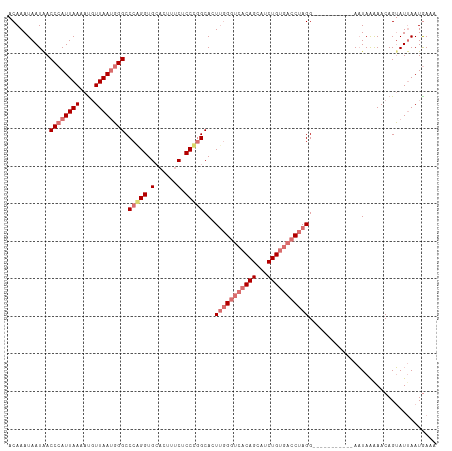
| Location | 799,909 – 800,023 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.60 |
| Shannon entropy | 0.28201 |
| G+C content | 0.50949 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -28.23 |
| Energy contribution | -29.35 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.677869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 799909 114 + 23011544 AACAGAUGCUGUGACCCAACUGCCGGGAGAAAGUGCACCUGGGCCCAUUAACAUUUUAAUGGGUUAUUAUUUGUCUCUGUGCAUCUGCAUCUGCUGGCGUUUUCGGGAGUUUCG ....((.(((....((((((.(((((.(((.(((((((..(((((((((((....)))))))))..........))..))))).))...))).))))))))...)))))).)). ( -35.60, z-score = -1.22, R) >droYak2.chr2L 782667 98 + 22324452 CACAGAUGCUGGG--CGGAGUGCAAGGUGGU----CACCUGGGCCGCUUAAGAUUUUAAUGGGUUAUUAUUUGUCUCCGUGCAUCUGCAUCUGGUGGCGCUUUG---------- ..(((((((((.(--(((((.((((((((((----(.....))))))))..(((((....)))))......))))))))).))...)))))))...........---------- ( -34.90, z-score = -1.62, R) >droSec1.super_14 769840 114 + 2068291 CACAGAUGCUGUGACCCAACUGCCGGGCGAAAGUGCACCCGGGCCCAUUAACAUUUUAAUGGGUCAUUAUUUGUCUCUGUGCAUCUGCAUCUGUCGGCGUUUUCGGGAGUUUCG ..(((((((.(.(((........((((.(......).))))((((((((((....)))))))))).......))).)...)))))))...((.((((.....)))).))..... ( -37.20, z-score = -1.30, R) >droSim1.chr2L 801314 114 + 22036055 CACAGAUGCUGUGACCCAAGUGCCGGGAGAAAGUGCACCCGGGCCCAUUAACGUUUUAAUGGGUCAUUAUUUGUCUCUGUGCAUCUGCAUCUGUCGGCGUUUUCGGGAGUUUCG (((((...))))).((((((((((((.(((.(((((((...((((((((((....)))))))))).............))))).))...))).)))))))))..)))....... ( -39.79, z-score = -1.90, R) >consensus CACAGAUGCUGUGACCCAACUGCCGGGAGAAAGUGCACCCGGGCCCAUUAACAUUUUAAUGGGUCAUUAUUUGUCUCUGUGCAUCUGCAUCUGUCGGCGUUUUCGGGAGUUUCG ..(((((((...((..(((((((((((.(......).)))))(((((((((....)))))))))...))))))..))...)))))))........................... (-28.23 = -29.35 + 1.12)

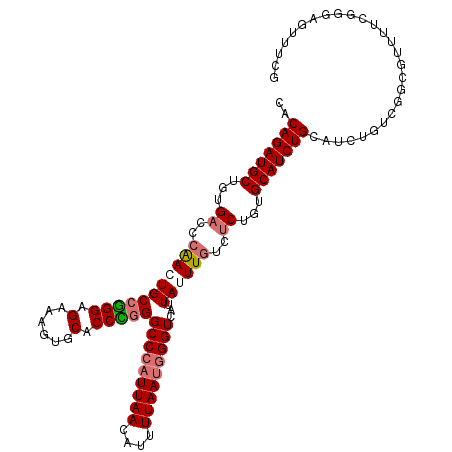
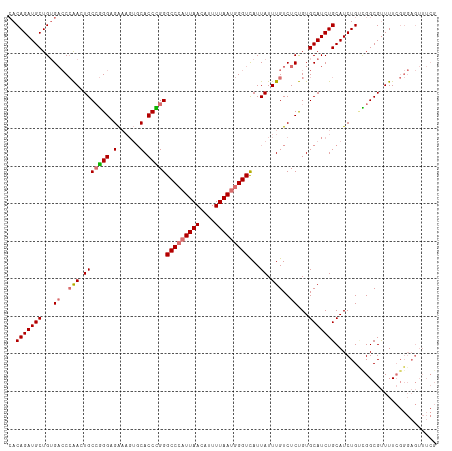
| Location | 799,909 – 800,023 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.60 |
| Shannon entropy | 0.28201 |
| G+C content | 0.50949 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -26.60 |
| Energy contribution | -28.47 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 799909 114 - 23011544 CGAAACUCCCGAAAACGCCAGCAGAUGCAGAUGCACAGAGACAAAUAAUAACCCAUUAAAAUGUUAAUGGGCCCAGGUGCACUUUCUCCCGGCAGUUGGGUCACAGCAUCUGUU ...................(((((((((.......................((((((((....))))))))(((((.(((.(........)))).))))).....))))))))) ( -33.20, z-score = -2.01, R) >droYak2.chr2L 782667 98 - 22324452 ----------CAAAGCGCCACCAGAUGCAGAUGCACGGAGACAAAUAAUAACCCAUUAAAAUCUUAAGCGGCCCAGGUG----ACCACCUUGCACUCCG--CCCAGCAUCUGUG ----------................((((((((..(....).........................((((.(.((((.----...)))).)....)))--)...)))))))). ( -24.10, z-score = -1.72, R) >droSec1.super_14 769840 114 - 2068291 CGAAACUCCCGAAAACGCCGACAGAUGCAGAUGCACAGAGACAAAUAAUGACCCAUUAAAAUGUUAAUGGGCCCGGGUGCACUUUCGCCCGGCAGUUGGGUCACAGCAUCUGUG ..........................((((((((...((..(((....((.((((((((....)))))))).(((((((......))))))))).)))..))...)))))))). ( -36.90, z-score = -2.18, R) >droSim1.chr2L 801314 114 - 22036055 CGAAACUCCCGAAAACGCCGACAGAUGCAGAUGCACAGAGACAAAUAAUGACCCAUUAAAACGUUAAUGGGCCCGGGUGCACUUUCUCCCGGCACUUGGGUCACAGCAUCUGUG ..........................((((((((.................((((((((....))))))))(((((((((.(........)))))))))).....)))))))). ( -36.30, z-score = -2.58, R) >consensus CGAAACUCCCGAAAACGCCAACAGAUGCAGAUGCACAGAGACAAAUAAUAACCCAUUAAAAUGUUAAUGGGCCCAGGUGCACUUUCUCCCGGCACUUGGGUCACAGCAUCUGUG ..........................((((((((.................((((((((....))))))))(((((((((...........))))))))).....)))))))). (-26.60 = -28.47 + 1.87)

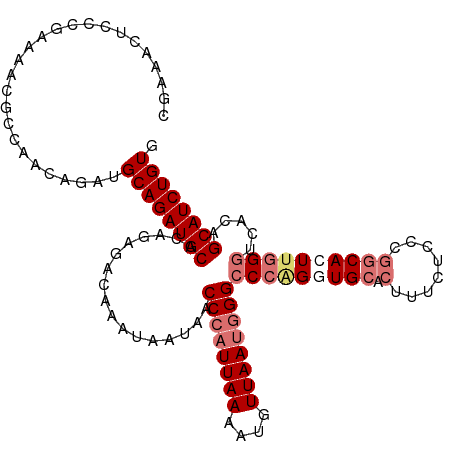
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:58 2011