
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,332,558 – 22,332,631 |
| Length | 73 |
| Max. P | 0.992897 |

| Location | 22,332,558 – 22,332,631 |
|---|---|
| Length | 73 |
| Sequences | 8 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 92.74 |
| Shannon entropy | 0.13207 |
| G+C content | 0.39671 |
| Mean single sequence MFE | -22.31 |
| Consensus MFE | -20.01 |
| Energy contribution | -20.51 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.991043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22332558 73 + 27905053 UUAAUUGCCACAUUAUGUGGCAGUGUUCCAAGUUGAUGCGUCCAACUUGGC--------AUAUUUCAAUUGUUAAUCCAGC ...((((((((.....))))))))((.((((((((.......)))))))).--------)).................... ( -21.70, z-score = -2.97, R) >droPer1.super_0 1718368 81 + 11822988 UUAAUUGCCACAUUAUGUGGCACUGUUCCAUGUUGAUGCAUUCAACUUGGCGGCAUAGCAUAUUUCAAUUGUUAAUCCAGC .....((((((.....)))))).(((((((.(((((.....))))).))).))))(((((.........)))))....... ( -21.70, z-score = -1.45, R) >dp4.chr2 7885592 81 + 30794189 UUAAUUGCCACAUUAUGUGGCACUGUUCCAAGUUGAUGCAUUCAACUUGGCGGCAUAGCAUAUUUCAAUUGUUAAUCCAGC .....((((((.....)))))).(((((((((((((.....))))))))).))))(((((.........)))))....... ( -25.80, z-score = -3.19, R) >droAna3.scaffold_13340 1380591 73 - 23697760 UUAAUUGCCACAUUAUGUGGCUGUGUUCCAAGUUGAUGCGUUCAACUUGGC--------AUAUUUCAAUUGUUAAUCCAGC .((((((((((.....))))).((((.(((((((((.....))))))))).--------))))...))))).......... ( -22.50, z-score = -3.29, R) >droEre2.scaffold_4820 4759850 73 - 10470090 UUAAUUGCCACAUUAUGUGGCAGUGUUCCAAGUUGAUGCGUCCAACUUGGC--------AUAUUUCAAUUGUUAAUCCAGC ...((((((((.....))))))))((.((((((((.......)))))))).--------)).................... ( -21.70, z-score = -2.97, R) >droYak2.chr3R 4678312 73 - 28832112 GUAAUUGCCACAUUAUGUGGCAGUGUUCCAAGUUGAUGCGUCCAACUUGGC--------AUAUUUCAAUUGUUAAUCCAGC ...((((((((.....))))))))((.((((((((.......)))))))).--------)).................... ( -21.70, z-score = -2.84, R) >droSec1.super_13 1124334 73 + 2104621 UUAAUUGCCACAUUAUGUGGCAGUGUUCCAAGUUGAUGCGUCCAACUUGGC--------AUAUUUCAAUUGUUAAUCCAGC ...((((((((.....))))))))((.((((((((.......)))))))).--------)).................... ( -21.70, z-score = -2.97, R) >droSim1.chr3R 22171246 73 + 27517382 UUAAUUGCCACAUUAUGUGGCAGUGUUCCAAGUUGAUGCGUCCAACUUGGC--------AUAUUUCAAUUGUUAAUCCAGC ...((((((((.....))))))))((.((((((((.......)))))))).--------)).................... ( -21.70, z-score = -2.97, R) >consensus UUAAUUGCCACAUUAUGUGGCAGUGUUCCAAGUUGAUGCGUCCAACUUGGC________AUAUUUCAAUUGUUAAUCCAGC ...((((((((.....))))))))...((((((((.......))))))))............................... (-20.01 = -20.51 + 0.50)

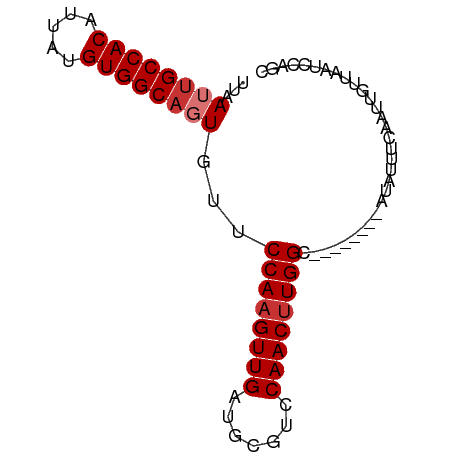
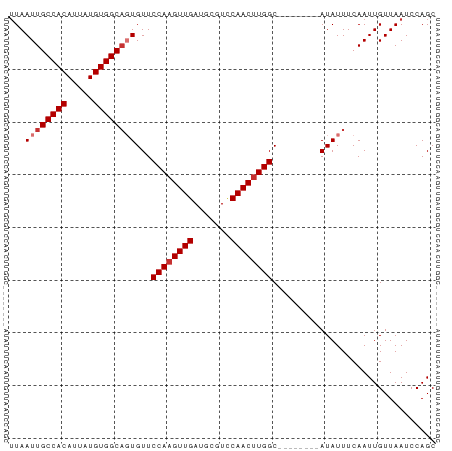
| Location | 22,332,558 – 22,332,631 |
|---|---|
| Length | 73 |
| Sequences | 8 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 92.74 |
| Shannon entropy | 0.13207 |
| G+C content | 0.39671 |
| Mean single sequence MFE | -20.69 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22332558 73 - 27905053 GCUGGAUUAACAAUUGAAAUAU--------GCCAAGUUGGACGCAUCAACUUGGAACACUGCCACAUAAUGUGGCAAUUAA ......................--------.(((((((((.....))))))))).....((((((.....))))))..... ( -20.30, z-score = -2.73, R) >droPer1.super_0 1718368 81 - 11822988 GCUGGAUUAACAAUUGAAAUAUGCUAUGCCGCCAAGUUGAAUGCAUCAACAUGGAACAGUGCCACAUAAUGUGGCAAUUAA ((((..................((...))..(((.(((((.....))))).)))..))))(((((.....)))))...... ( -19.40, z-score = -0.64, R) >dp4.chr2 7885592 81 - 30794189 GCUGGAUUAACAAUUGAAAUAUGCUAUGCCGCCAAGUUGAAUGCAUCAACUUGGAACAGUGCCACAUAAUGUGGCAAUUAA ((((..................((...))..(((((((((.....)))))))))..))))(((((.....)))))...... ( -23.50, z-score = -2.31, R) >droAna3.scaffold_13340 1380591 73 + 23697760 GCUGGAUUAACAAUUGAAAUAU--------GCCAAGUUGAACGCAUCAACUUGGAACACAGCCACAUAAUGUGGCAAUUAA ......................--------.(((((((((.....)))))))))......(((((.....)))))...... ( -21.10, z-score = -3.44, R) >droEre2.scaffold_4820 4759850 73 + 10470090 GCUGGAUUAACAAUUGAAAUAU--------GCCAAGUUGGACGCAUCAACUUGGAACACUGCCACAUAAUGUGGCAAUUAA ......................--------.(((((((((.....))))))))).....((((((.....))))))..... ( -20.30, z-score = -2.73, R) >droYak2.chr3R 4678312 73 + 28832112 GCUGGAUUAACAAUUGAAAUAU--------GCCAAGUUGGACGCAUCAACUUGGAACACUGCCACAUAAUGUGGCAAUUAC ......................--------.(((((((((.....))))))))).....((((((.....))))))..... ( -20.30, z-score = -2.90, R) >droSec1.super_13 1124334 73 - 2104621 GCUGGAUUAACAAUUGAAAUAU--------GCCAAGUUGGACGCAUCAACUUGGAACACUGCCACAUAAUGUGGCAAUUAA ......................--------.(((((((((.....))))))))).....((((((.....))))))..... ( -20.30, z-score = -2.73, R) >droSim1.chr3R 22171246 73 - 27517382 GCUGGAUUAACAAUUGAAAUAU--------GCCAAGUUGGACGCAUCAACUUGGAACACUGCCACAUAAUGUGGCAAUUAA ......................--------.(((((((((.....))))))))).....((((((.....))))))..... ( -20.30, z-score = -2.73, R) >consensus GCUGGAUUAACAAUUGAAAUAU________GCCAAGUUGGACGCAUCAACUUGGAACACUGCCACAUAAUGUGGCAAUUAA ...............................(((((((((.....))))))))).....((((((.....))))))..... (-20.08 = -20.10 + 0.02)
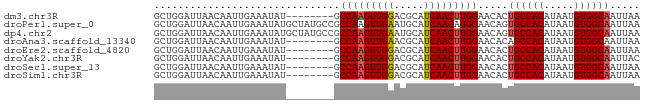
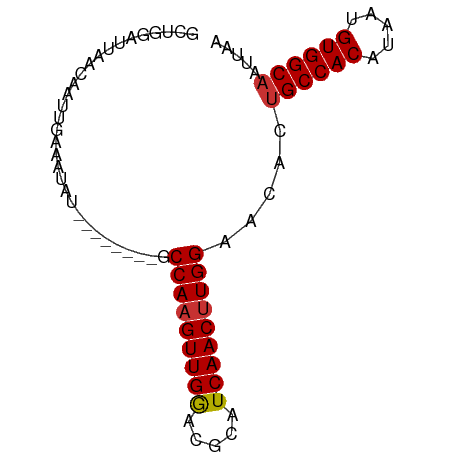
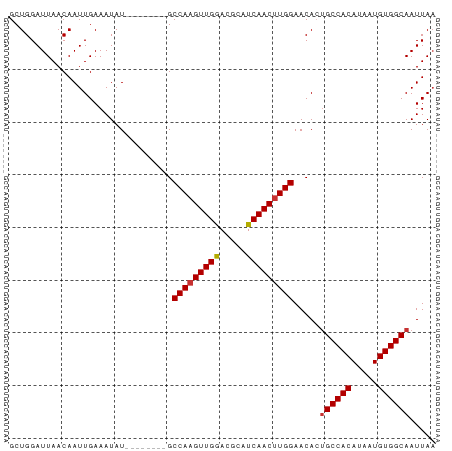
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:52 2011