| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,325,180 – 22,325,275 |
| Length | 95 |
| Max. P | 0.985161 |

| Location | 22,325,180 – 22,325,275 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 85.99 |
| Shannon entropy | 0.26391 |
| G+C content | 0.44642 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -22.00 |
| Energy contribution | -23.12 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
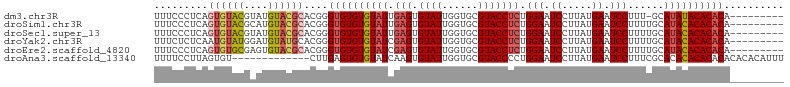
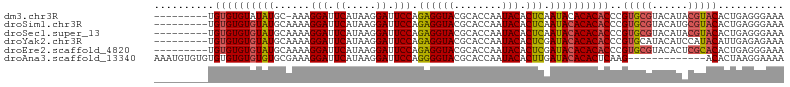
>dm3.chr3R 22325180 95 + 27905053 UUUCCCUCAGUGUACGUAUGUACGCACGGGUGUGUGUAUUGAGUGUAUUGGUGCGUACCUCUGGAAUCCUUAUGAAUCCUUU-GCAUAUACACACA--------- ...(((...((((((....))))))..)))(((((((((...(((((..((..((((.....((...)).))))...))..)-)))))))))))))--------- ( -28.30, z-score = -1.39, R) >droSim1.chr3R 22163903 96 + 27517382 UUUCCCUCAGUGUACGCAUGUACGCACGGGUGUGUGUAUUGAGUGUAUUGGUGCGUACCUCUGGAAUCCUUAUGAAUCCUUUUGCAUACACACACA--------- .........(((((.(((.((((((((.((((..(.......)..)))).))))))))....(((.((.....)).)))...))).))))).....--------- ( -30.10, z-score = -1.70, R) >droSec1.super_13 1117043 96 + 2104621 UUUCCCUCAGUGUACGUAUGUACGCACGGGUGUGUGUAUUGAGUGUAUUGGUGCGUACCUCUGGAAUCCUUAUGAAUCCUUUUGCAUACACACACA--------- ....((...((((((....))))))..))((((((((((.(((.((((......))))))).(((.((.....)).)))......)))))))))).--------- ( -29.80, z-score = -1.75, R) >droYak2.chr3R 4670628 96 - 28832112 UUUCUCUCAAUGUAUGGAUGUAUGCACGGGUGUGUGUAUCGAGUGUAUUGGUGCGUACCUCUGGAAUCCUUAUGAAUCCUUUUGCAUACACACACA--------- ..............((..((((((((.(((.(((((((((((.....)))))))))))))).(((.((.....)).)))...))))))))..))..--------- ( -27.70, z-score = -1.40, R) >droEre2.scaffold_4820 4751888 96 - 10470090 UUUCCCUCAGUGUGCGAGUGUACGCACGGGUGUGUGUAUCGAGUGUAUUGGUGCGUACCUCUGGAAUCCUUAUGAAUCCUUUUGCAUACACACACA--------- .........(((((((((.((((((((.((((..(.......)..)))).))))))))))).(((.((.....)).)))....)))))).......--------- ( -34.10, z-score = -2.62, R) >droAna3.scaffold_13340 1371390 92 - 23697760 UUUUCCUUAGUGU-------------CUUGAGUGUGUAUCAAGUGUAUUGGUGCGUACCCCUGGAAUCCUUAUGAAUCCUUUCGCACACACACACACACACAUUU .........((((-------------..((.((((((..(((.....)))(((((.......(((.((.....)).)))...))))))))))).)).)))).... ( -23.30, z-score = -1.95, R) >consensus UUUCCCUCAGUGUACGUAUGUACGCACGGGUGUGUGUAUCGAGUGUAUUGGUGCGUACCUCUGGAAUCCUUAUGAAUCCUUUUGCAUACACACACA_________ .........((((((....))))))....((((((((((.(((.((((......))))))).(((.((.....)).)))......)))))))))).......... (-22.00 = -23.12 + 1.11)
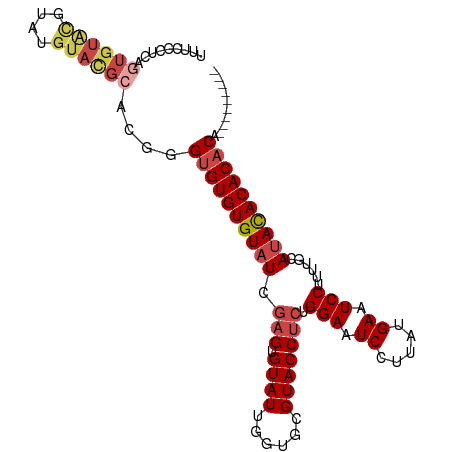
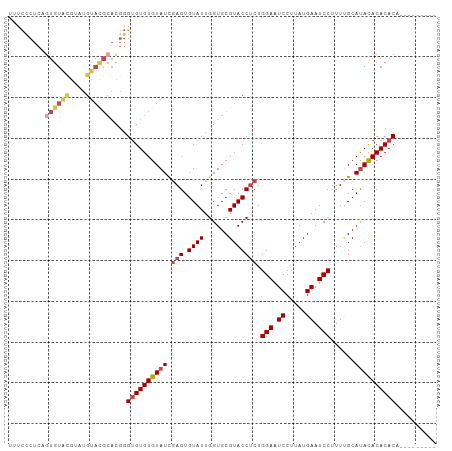
| Location | 22,325,180 – 22,325,275 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 85.99 |
| Shannon entropy | 0.26391 |
| G+C content | 0.44642 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -24.58 |
| Energy contribution | -25.02 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
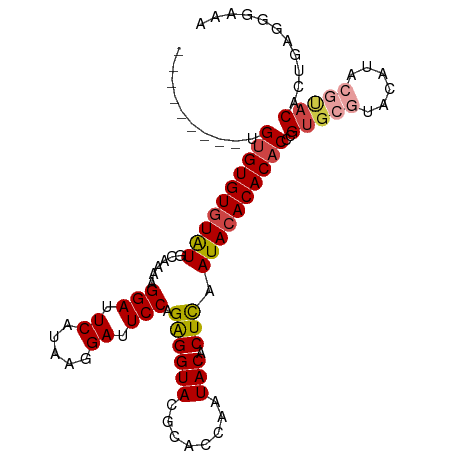
>dm3.chr3R 22325180 95 - 27905053 ---------UGUGUGUAUAUGC-AAAGGAUUCAUAAGGAUUCCAGAGGUACGCACCAAUACACUCAAUACACACACCCGUGCGUACAUACGUACACUGAGGGAAA ---------(((((((((....-...(((.((.....)).))).((((((........))).))).)))))))))(((..(.((((....)))).)...)))... ( -29.40, z-score = -3.02, R) >droSim1.chr3R 22163903 96 - 27517382 ---------UGUGUGUGUAUGCAAAAGGAUUCAUAAGGAUUCCAGAGGUACGCACCAAUACACUCAAUACACACACCCGUGCGUACAUGCGUACACUGAGGGAAA ---------.((((((((((......(((.((.....)).))).((((((........))).))).))))))))))..((((((....))))))........... ( -30.20, z-score = -2.31, R) >droSec1.super_13 1117043 96 - 2104621 ---------UGUGUGUGUAUGCAAAAGGAUUCAUAAGGAUUCCAGAGGUACGCACCAAUACACUCAAUACACACACCCGUGCGUACAUACGUACACUGAGGGAAA ---------.((((((((((......(((.((.....)).))).((((((........))).))).))))))))))..((((((....))))))........... ( -29.90, z-score = -2.60, R) >droYak2.chr3R 4670628 96 + 28832112 ---------UGUGUGUGUAUGCAAAAGGAUUCAUAAGGAUUCCAGAGGUACGCACCAAUACACUCGAUACACACACCCGUGCAUACAUCCAUACAUUGAGAGAAA ---------((((((((((((((...(((.((.....)).))).((((((........))).)))..............))))))))..)))))).......... ( -24.30, z-score = -1.91, R) >droEre2.scaffold_4820 4751888 96 + 10470090 ---------UGUGUGUGUAUGCAAAAGGAUUCAUAAGGAUUCCAGAGGUACGCACCAAUACACUCGAUACACACACCCGUGCGUACACUCGCACACUGAGGGAAA ---------.((((((((((.(....(((.((.....)).))).((((((........))).))))))))))))))..(((((......)))))........... ( -31.00, z-score = -2.61, R) >droAna3.scaffold_13340 1371390 92 + 23697760 AAAUGUGUGUGUGUGUGUGUGCGAAAGGAUUCAUAAGGAUUCCAGGGGUACGCACCAAUACACUUGAUACACACUCAAG-------------ACACUAAGGAAAA ...((.((((((((((((((((....(((.((.....)).)))....))))))))(((.....))))))))))).))..-------------............. ( -26.40, z-score = -1.68, R) >consensus _________UGUGUGUGUAUGCAAAAGGAUUCAUAAGGAUUCCAGAGGUACGCACCAAUACACUCAAUACACACACCCGUGCGUACAUACGUACACUGAGGGAAA ..........((((((((((......(((.((.....)).))).((((((........))).))).))))))))))..(((((......)))))........... (-24.58 = -25.02 + 0.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:50 2011