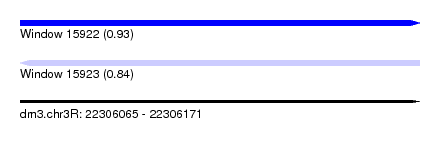
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,306,065 – 22,306,171 |
| Length | 106 |
| Max. P | 0.933636 |

| Location | 22,306,065 – 22,306,171 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.57 |
| Shannon entropy | 0.38073 |
| G+C content | 0.58066 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -21.63 |
| Energy contribution | -24.83 |
| Covariance contribution | 3.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
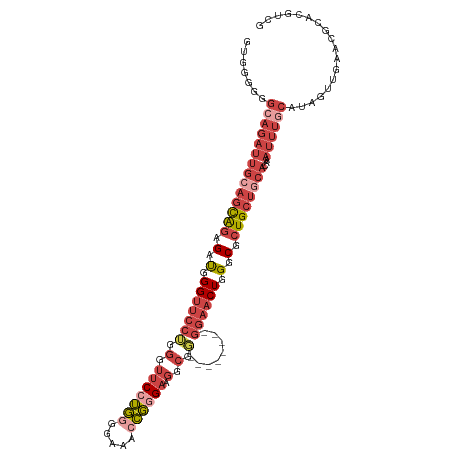
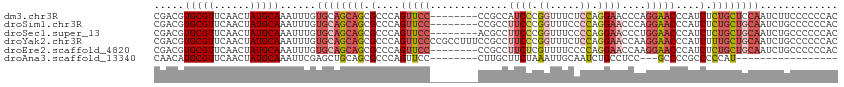
>dm3.chr3R 22306065 106 + 27905053 GUGGGGGGAAGAUUGGAGCAGAGAUGGGUUCCUGGGUUCCUGGAGAAACCGGGAUGGCGG--------GGAACUGGGCGCUGCUGCACAAAUUUGCAUAGUUGAACGCACGUCG (((.(.....(((((.(((((...(.(((((((.(.(((((((.....)))))).).).)--------)))))).)...)))))(((......))).)))))...).))).... ( -38.20, z-score = -1.98, R) >droSim1.chr3R 22144871 106 + 27517382 GUGGGGGGCAGAUUGCAGCAGAGAUGGGUUCCUGGGUUCCUGGGGAAACCGGGAAGGCGG--------GGAACUGGGCGCUGCUGCACAAAUUUGCAUAGUUGAACGCACGUCG (((.(..((((((((((((((...(.(((((((.(.(((((((.....)))))))..).)--------)))))).)...))))))))...)))))).........).))).... ( -47.00, z-score = -3.87, R) >droSec1.super_13 1097485 106 + 2104621 GUGGGGGGCAGAUUGCAGCAGAGAUGGGUUCCAGGGUUCCUGGGGAAACCGGGAAGGCGU--------GGAACUGGGCGCUGCUGCACAAAUUUGCAUAGUUGAACGCACGUCG (((.(..((((((((((((((...(.(((((((.(.(((((((.....)))))))..).)--------)))))).)...))))))))...)))))).........).))).... ( -47.00, z-score = -4.06, R) >droYak2.chr3R 4649924 114 - 28832112 GUGGGGGGCAGAUUGCAGCAAAGAUGGGUUCCUUGGUUCCUGGAGAAACCGGGAAGGCGGAAAGGCGGGGAACUGGGCGCUGCUGCACAAAUUUGCAUAGUUGAACGCACGUCG (((.(..(((((((((((((..(.(.((((((((..(((((((.....))))))).((......)))))))))).).)..)))))))...)))))).........).))).... ( -42.30, z-score = -1.97, R) >droEre2.scaffold_4820 4732011 106 - 10470090 GUGGGGGGCAGAUUGCAGCAGAGAUGGGUUCCUUGGUUCCUGGGGAAAACGAGAAGGCGG--------GGAACUGGGCGCUGCUGCACAAAUUUGCAUAGUUGAACGCACGUCG (((.(..((((((((((((((...(.(((((((((.((.((.(......).)).)).)))--------)))))).)...))))))))...)))))).........).))).... ( -41.30, z-score = -2.91, R) >droAna3.scaffold_13340 1350415 86 - 23697760 -----------------AUGGGGGCGGGGC---GGAGGCAGAUUGCAAUUUAGAAGCAAG--------GGAACUGGGCGCUGCAGCUCGAAUUUGCAUAGUUGAACGCAUGUUG -----------------.....((((..((---(....(((..(((((....((.((.((--------....))..))((....))))....)))))...)))..))).)))). ( -20.60, z-score = 0.64, R) >consensus GUGGGGGGCAGAUUGCAGCAGAGAUGGGUUCCUGGGUUCCUGGGGAAACCGGGAAGGCGG________GGAACUGGGCGCUGCUGCACAAAUUUGCAUAGUUGAACGCACGUCG .......((((((((((((((.(.(.((((((....(((((((.....))))))).............)))))).).).))))))))...)))))).................. (-21.63 = -24.83 + 3.20)

| Location | 22,306,065 – 22,306,171 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.57 |
| Shannon entropy | 0.38073 |
| G+C content | 0.58066 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -16.81 |
| Energy contribution | -16.95 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22306065 106 - 27905053 CGACGUGCGUUCAACUAUGCAAAUUUGUGCAGCAGCGCCCAGUUCC--------CCGCCAUCCCGGUUUCUCCAGGAACCCAGGAACCCAUCUCUGCUCCAAUCUUCCCCCCAC (((..(((((......)))))...)))((.(((((.(....(((((--------......(((.((.....)).))).....)))))....).))))).))............. ( -22.80, z-score = -1.16, R) >droSim1.chr3R 22144871 106 - 27517382 CGACGUGCGUUCAACUAUGCAAAUUUGUGCAGCAGCGCCCAGUUCC--------CCGCCUUCCCGGUUUCCCCAGGAACCCAGGAACCCAUCUCUGCUGCAAUCUGCCCCCCAC (((..(((((......)))))...)))((((((((.(....(((((--------..(..((((.((.....)).))))..).)))))....).))))))))............. ( -30.30, z-score = -2.67, R) >droSec1.super_13 1097485 106 - 2104621 CGACGUGCGUUCAACUAUGCAAAUUUGUGCAGCAGCGCCCAGUUCC--------ACGCCUUCCCGGUUUCCCCAGGAACCCUGGAACCCAUCUCUGCUGCAAUCUGCCCCCCAC (((..(((((......)))))...)))((((((((.(....(((((--------(.(..((((.((.....)).))))..)))))))....).))))))))............. ( -32.40, z-score = -3.16, R) >droYak2.chr3R 4649924 114 + 28832112 CGACGUGCGUUCAACUAUGCAAAUUUGUGCAGCAGCGCCCAGUUCCCCGCCUUUCCGCCUUCCCGGUUUCUCCAGGAACCAAGGAACCCAUCUUUGCUGCAAUCUGCCCCCCAC (((..(((((......)))))...)))((((((((.(....(((((..((......))......((((((....))))))..)))))....).))))))))............. ( -29.80, z-score = -1.95, R) >droEre2.scaffold_4820 4732011 106 + 10470090 CGACGUGCGUUCAACUAUGCAAAUUUGUGCAGCAGCGCCCAGUUCC--------CCGCCUUCUCGUUUUCCCCAGGAACCAAGGAACCCAUCUCUGCUGCAAUCUGCCCCCCAC (((..(((((......)))))...)))((((((((.(....(((((--------..(..((((.(......).))))..)..)))))....).))))))))............. ( -24.90, z-score = -1.50, R) >droAna3.scaffold_13340 1350415 86 + 23697760 CAACAUGCGUUCAACUAUGCAAAUUCGAGCUGCAGCGCCCAGUUCC--------CUUGCUUCUAAAUUGCAAUCUGCCUCC---GCCCCGCCCCCAU----------------- ......(((.........(((.....((((((.......)))))).--------.((((.........))))..)))....---....)))......----------------- ( -12.13, z-score = 0.23, R) >consensus CGACGUGCGUUCAACUAUGCAAAUUUGUGCAGCAGCGCCCAGUUCC________CCGCCUUCCCGGUUUCCCCAGGAACCCAGGAACCCAUCUCUGCUGCAAUCUGCCCCCCAC .....(((((......)))))......((((((((.(....(((((.........((......))..((((...))))....)))))....).))))))))............. (-16.81 = -16.95 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:47 2011