
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,305,213 – 22,305,322 |
| Length | 109 |
| Max. P | 0.597948 |

| Location | 22,305,213 – 22,305,322 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.70 |
| Shannon entropy | 0.34257 |
| G+C content | 0.43918 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -24.19 |
| Energy contribution | -26.31 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22305213 109 + 27905053 UAUUUUUCCAAGUGCAAUUGCAGUAGCAGUUGCAUGUGCAACCAGGGCGUAAUUGUUAUAGCGUAAUUGUUUCAAUGGCGGGGCGUUUAAUUGCGCUUUGGCAACUACU ........((.((((((((((....)))))))))).))...((((((((((((((....(((((...((((.....))))..)))))))))))))))))))........ ( -37.50, z-score = -2.37, R) >droEre2.scaffold_4820 4731132 109 - 10470090 UGCUUUUCCAAGUGCAAGUGCAGUAGCAGUUGCAUGUGCAACCAGGGCGUAAUUGUUAUAGCGUAAUUGUUUCAAUGGCGGGGCGUUUAAUUGCGCUUUGGCAACUACU .(((((..((.((((((.(((....))).)))))).)).....)))))(((.((((((.(((((((((((..(......)..))....))))))))).)))))).))). ( -37.10, z-score = -1.36, R) >droYak2.chr3R 4649058 109 - 28832112 UAUUUUUGCAAGUGCAAGUGCAGUAGCAGUUGCAUGUGCAACCAGGGCGUAAUUGUUAUAGCGUAAUUGUUUCAAUGGCGGGGCGUUUAAUUGCGCUUUGGCAACUACU .....(((((.((((((.(((....))).)))))).)))))((((((((((((((....(((((...((((.....))))..)))))))))))))))))))........ ( -41.80, z-score = -3.27, R) >droSec1.super_13 1096633 109 + 2104621 UAUUUUUUCAAGUGCAAAUGCAGUAGCAGUUGCAUGUGCAACCAGGGCGUAAUUGUUAUAGCGUAAUUGUUUCAAUGGCGGGGCGUUUAAUUGCGCUUUGGCAACUACU ............((((.((((((......)))))).)))).((((((((((((((....(((((...((((.....))))..)))))))))))))))))))........ ( -33.80, z-score = -1.43, R) >droSim1.chr3R 22144017 109 + 27517382 UAUUUUUCCAAGUGCAAAUGCAGUAGCAGUUGCAUGUGCAACCAGGGCGUAAUUGUUAUAGCGUAAUUGUUUCAAUGGCGGGGCGUUUAAUUGCGCUUUGGCAACUACU ......(((...((((.((((((......)))))).))))....))).(((.((((((.(((((((((((..(......)..))....))))))))).)))))).))). ( -34.00, z-score = -1.19, R) >droWil1.scaffold_181089 2447109 105 - 12369635 UGUGUUACGGAUUGCAUGUGCAG--GCGGCGGCACAGGCA--CAGGGCGUAAUUGUUAUGGCGUAAUUGUUUCAAUGGCGGGGCGUUUAAUUGCGCUUCGGAAACUACU (((((..(.(.((((....))))--.).)..))))).((.--((..(((....)))..))))(((...(((((.....((((((((......)))))))))))))))). ( -32.20, z-score = -0.46, R) >droMoj3.scaffold_6540 18811723 85 + 34148556 ------------------------UGGCUUUGCAUGUGCAACCAAAACGUAAUUGUUAUGGCAUAAUUAUUUCAAUGGCAGGGCGUUAAAUUGCACUUUAGAAACUGCU ------------------------.((((((....((((((....(((((..(((((((...............))))))).)))))...))))))....)))...))) ( -16.16, z-score = 0.98, R) >consensus UAUUUUUCCAAGUGCAAGUGCAGUAGCAGUUGCAUGUGCAACCAGGGCGUAAUUGUUAUAGCGUAAUUGUUUCAAUGGCGGGGCGUUUAAUUGCGCUUUGGCAACUACU ............((((.((((((......)))))).)))).((((((((((((((....(((((...((((.....))))..)))))))))))))))))))........ (-24.19 = -26.31 + 2.13)

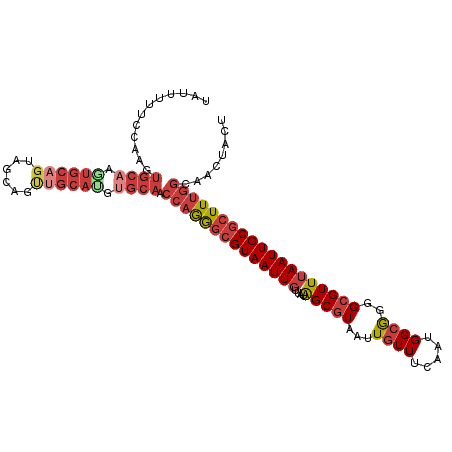
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:45 2011