
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,286,505 – 22,286,595 |
| Length | 90 |
| Max. P | 0.604761 |

| Location | 22,286,505 – 22,286,595 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 61.37 |
| Shannon entropy | 0.74523 |
| G+C content | 0.55786 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -9.82 |
| Energy contribution | -9.28 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.604761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
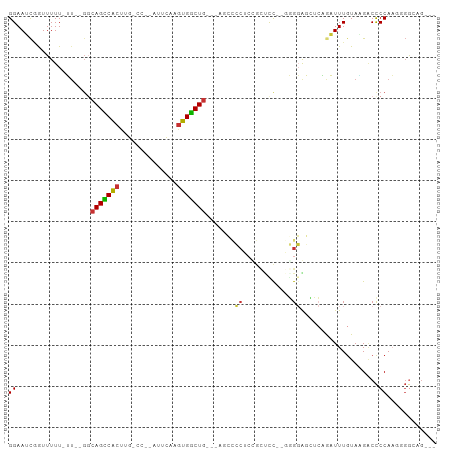
>dm3.chr3R 22286505 90 + 27905053 GGAAUCGAUUUUU-UU--GGCAGUCACUUGCCAGCAUUCAAGUGGCUGCG-----CCGCUGCUCC---GGGAGCUCAGAUUUGUAAGACCCCAAGGGGCAG--- ((...(((.....-))--)(((((((((((........))))))))))).-----)).(((((((---(((..((..........))..)))..)))))))--- ( -33.40, z-score = -1.39, R) >droEre2.scaffold_4820 4710957 82 - 10470090 GGAAUCGGUUUUU-U--GGCCAGCCA-------------AAGUGGCUGCUGGGCUCCUCCGCCUU--GGGGGGCUCGGAUUUGUAGGACCCCAAUGGGCG---- (((...((...((-.--.((.((((.-------------....))))))..))..)))))((((.--..((((..(.........)..))))...)))).---- ( -32.50, z-score = 0.18, R) >droYak2.chr3R 4630108 100 - 28832112 GGAAUCGGUUUUU-UUUGCCCAGUCACUUGGCCACAUUCAAGUGACUGCUGAGCCCCUCUGCCUUUGGGGGAGCUCAGAUUUGUAUGGCCCCAAUGGGCGG--- .((((((((....-...)))(((((((((((......))))))))))).((((((((((.......))))).)))))))))).....((((....))))..--- ( -43.30, z-score = -2.87, R) >droSec1.super_13 1077726 79 + 2104621 GGAAUCGGUUUUU-UU--GGCAGUCACUUGCCCGCAUUCAAGUGGCU----------------CC---GGGAGCUCAGAUUUGUAAGACCCCAAGGGGCAG--- .(((((.(..(((-((--((.(((((((((........)))))))))----------------))---)))))..).)))))....(.(((....))))..--- ( -27.10, z-score = -1.23, R) >droSim1.chr3R 22125313 95 + 27517382 GGAAUCCGUUUUU-UU--GGCAGUCACUUGCCCGCAUUCAAGUGGCUGCUGAGCCCCUCUGCUCC---GGGAGCUCAGAUUUGUAAGACCCCAAGGGGCAG--- .(((((.......-..--((((((((((((........))))))))))))(((((((........---))).)))).)))))....(.(((....))))..--- ( -38.00, z-score = -2.08, R) >droPer1.super_0 1666982 95 + 11822988 GGAAACAAUUUUUUGUGUGGCAGCCCCCC------AUUGAUGGGGCUG---AAGCGCCCCGCUCCAGGUGCGCAAAUAUUUUGUAUGACCCCAUCUGAAGGGCG (....)........((((..(((((((..------......)))))))---..))))..(((((((((((.(...((((...))))...).))))))..))))) ( -33.30, z-score = -1.20, R) >dp4.chr2 7841760 95 + 30794189 GGAAACAAUUUUU-GUGUGGCAGCCCCCCC-----AUUGAUGGGGCUG---AAGCGCCCAGCUCCAGGUGCGCGAAUAUUUUAUAUGACCCCAUCUGAAGGGAG (....).....(.-.((((((((((((...-----......)))))))---..((((((.......)).)))).......)))))..).(((.......))).. ( -30.00, z-score = -0.72, R) >droAna3.scaffold_13340 1330266 91 - 23697760 GGAAUUGGUUUUU-GU--GGCAGCCACUUGGCC----UAAAGUGGCUG---GGGCCCUCCGUUUCCGGAGGGCAACUAUUUUGUACGCCUCCAAAUUGGAG--- .(((.(((((...-..--..(((((((((....----..)))))))))---..((((((((....))))))))))))).)))......((((.....))))--- ( -40.40, z-score = -3.12, R) >consensus GGAAUCGGUUUUU_UU__GGCAGCCACUUG_CC__AUUCAAGUGGCUG___AGCCCCUCCGCUCC__GGGGAGCUCAGAUUUGUAAGACCCCAAGGGGCAG___ ((..................(((((((..............))))))).......((............))...................))............ ( -9.82 = -9.28 + -0.55)

| Location | 22,286,505 – 22,286,595 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 61.37 |
| Shannon entropy | 0.74523 |
| G+C content | 0.55786 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -9.38 |
| Energy contribution | -9.60 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
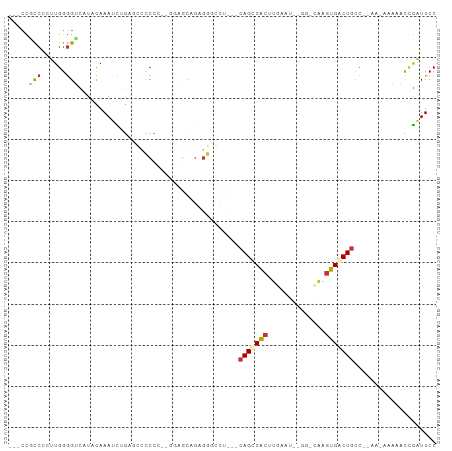
>dm3.chr3R 22286505 90 - 27905053 ---CUGCCCCUUGGGGUCUUACAAAUCUGAGCUCCC---GGAGCAGCGG-----CGCAGCCACUUGAAUGCUGGCAAGUGACUGCC--AA-AAAAAUCGAUUCC ---((((..(((.(((.((((......))))..)))---.)))..))))-----.((((.((((((........)))))).)))).--..-............. ( -28.60, z-score = -0.86, R) >droEre2.scaffold_4820 4710957 82 + 10470090 ----CGCCCAUUGGGGUCCUACAAAUCCGAGCCCCCC--AAGGCGGAGGAGCCCAGCAGCCACUU-------------UGGCUGGCC--A-AAAAACCGAUUCC ----.(((..(((((.(((......(((..(((....--..))))))))).))))).((((....-------------.))))))).--.-............. ( -26.10, z-score = -0.05, R) >droYak2.chr3R 4630108 100 + 28832112 ---CCGCCCAUUGGGGCCAUACAAAUCUGAGCUCCCCCAAAGGCAGAGGGGCUCAGCAGUCACUUGAAUGUGGCCAAGUGACUGGGCAAA-AAAAACCGAUUCC ---...(((...)))(((........(((((((((((....))....)))))))))((((((((((........)))))))))))))...-............. ( -38.10, z-score = -2.16, R) >droSec1.super_13 1077726 79 - 2104621 ---CUGCCCCUUGGGGUCUUACAAAUCUGAGCUCCC---GG----------------AGCCACUUGAAUGCGGGCAAGUGACUGCC--AA-AAAAACCGAUUCC ---.........(((..((((......))))..)))---((----------------((.((((((........)))))).)).))--..-............. ( -18.70, z-score = 0.45, R) >droSim1.chr3R 22125313 95 - 27517382 ---CUGCCCCUUGGGGUCUUACAAAUCUGAGCUCCC---GGAGCAGAGGGGCUCAGCAGCCACUUGAAUGCGGGCAAGUGACUGCC--AA-AAAAACGGAUUCC ---..((((....))))......(((((((((((((---......).))))))).((((.((((((........)))))).)))).--..-......))))).. ( -34.60, z-score = -1.10, R) >droPer1.super_0 1666982 95 - 11822988 CGCCCUUCAGAUGGGGUCAUACAAAAUAUUUGCGCACCUGGAGCGGGGCGCUU---CAGCCCCAUCAAU------GGGGGGCUGCCACACAAAAAAUUGUUUCC .((((((((((((((((..............((((.((((...))))))))..---..))))))))..)------)))))))...................... ( -36.07, z-score = -1.93, R) >dp4.chr2 7841760 95 - 30794189 CUCCCUUCAGAUGGGGUCAUAUAAAAUAUUCGCGCACCUGGAGCUGGGCGCUU---CAGCCCCAUCAAU-----GGGGGGGCUGCCACAC-AAAAAUUGUUUCC ((((((...((((((((.((((...))))..((((.((.......))))))..---..))))))))...-----))))))..........-............. ( -31.70, z-score = -0.88, R) >droAna3.scaffold_13340 1330266 91 + 23697760 ---CUCCAAUUUGGAGGCGUACAAAAUAGUUGCCCUCCGGAAACGGAGGGCCC---CAGCCACUUUA----GGCCAAGUGGCUGCC--AC-AAAAACCAAUUCC ---.(((.....)))(((.............((((((((....))))))))..---.((((((((..----....)))))))))))--..-............. ( -40.10, z-score = -4.83, R) >consensus ___CCGCCCCUUGGGGUCAUACAAAUCUGAGCCCCCC__GGAGCAGAGGGCCU___CAGCCACUUGAAU__GG_CAAGUGACUGCC__AA_AAAAACCGAUUCC .....(((......))).......................................(((((((.((........)).))))))).................... ( -9.38 = -9.60 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:43 2011