| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,268,921 – 22,269,024 |
| Length | 103 |
| Max. P | 0.951985 |

| Location | 22,268,921 – 22,269,024 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 69.23 |
| Shannon entropy | 0.58058 |
| G+C content | 0.36218 |
| Mean single sequence MFE | -18.37 |
| Consensus MFE | -6.20 |
| Energy contribution | -6.62 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
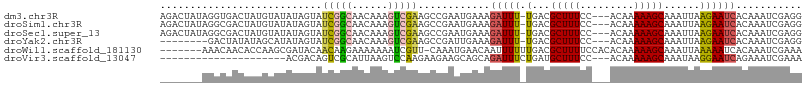
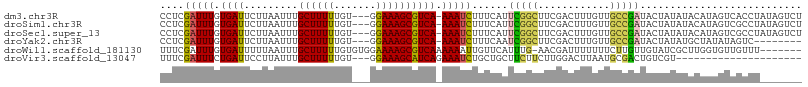
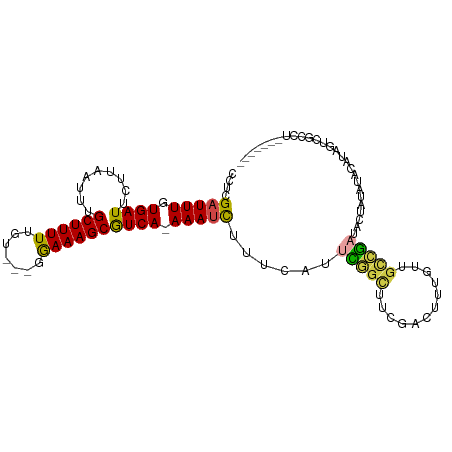
>dm3.chr3R 22268921 103 + 27905053 AGACUAUAGGUGACUAUGUAUAUAGUAUCGGCAACAAAGUCGAAGCCGAAUGAAAGAUUU-UGACGCUUUCC---ACAAAAAGCAAAUUAAGAAUCACAAAUCGAGG ....(((((....))))).........(((((..(......)..)))))......(((((-(...(((((..---....)))))......))))))........... ( -20.00, z-score = -1.50, R) >droSim1.chr3R 22107884 103 + 27517382 AGACUAUAGGCGACUAUGUAUAUAGUAUCGGCAACAAAGUCGAAGCCGAAUGAAAGAUUU-UGACGCUUUCC---ACAAAAAGCAAAUUAAGAAUCACAAAUCGAGG ........(((.(((((....))))).(((((......))))).)))........(((((-(...(((((..---....)))))......))))))........... ( -20.10, z-score = -1.53, R) >droSec1.super_13 1060297 103 + 2104621 AGACUAUAGGCGACUAUGUAUAUAGUAUCGGCAACAAAGUCGAAGCCGAAUGAAAGAUUU-UGACGCUUUCC---ACAAAAAGCAAAUUAAGAAUCACAAAUCGAGG ........(((.(((((....))))).(((((......))))).)))........(((((-(...(((((..---....)))))......))))))........... ( -20.10, z-score = -1.53, R) >droYak2.chr3R 4612338 95 - 28832112 --------GACUAUAUAGCAUAUAGUAUCGGCAACAAAGUCGAAGCCGAUUGAAAGAUUU-UGACGCUUUCC---ACAAAAAGCAAAUUAAGAAUCACAAAUCGAGG --------.(((((((...))))))).(((((......)))))...(((((....(((((-(...(((((..---....)))))......))))))...)))))... ( -19.80, z-score = -2.74, R) >droWil1.scaffold_181130 7128788 99 + 16660200 -------AAACAACACCAAGCGAUACAACAAGAAAAAAAUCGUU-CAAAUGAACAAUUUUUUGACGCUUUUCCACACAAAAAGCAAAUUAAAAAUCACAAAUCGAAA -------.............((((.........(((((((.(((-(....)))).)))))))...((((((.......))))))................))))... ( -14.30, z-score = -3.39, R) >droVir3.scaffold_13047 4810320 83 + 19223366 ---------------------ACGACAGUCGCAUUAAGUCCAAGAAGAAGCAGCAGAUUUCUGAUGCUUUCC---ACAAAAAGCAAAUAAGGAAUCAGAAAUCGAAA ---------------------.(((...((((......((......))....)).(((((((..((((((..---....))))))....))))))).))..)))... ( -15.90, z-score = -1.53, R) >consensus _______AGGCGACUAUGUAUAUAGUAUCGGCAACAAAGUCGAAGCCGAAUGAAAGAUUU_UGACGCUUUCC___ACAAAAAGCAAAUUAAGAAUCACAAAUCGAGG ...........................(((((......)))))............(((((.(((.(((((.........)))))...))).)))))........... ( -6.20 = -6.62 + 0.42)
| Location | 22,268,921 – 22,269,024 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.23 |
| Shannon entropy | 0.58058 |
| G+C content | 0.36218 |
| Mean single sequence MFE | -19.42 |
| Consensus MFE | -11.38 |
| Energy contribution | -11.17 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
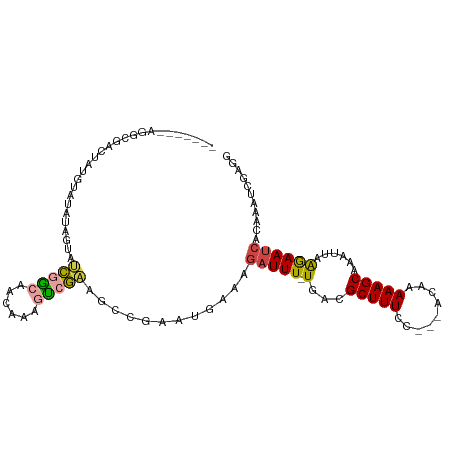
>dm3.chr3R 22268921 103 - 27905053 CCUCGAUUUGUGAUUCUUAAUUUGCUUUUUGU---GGAAAGCGUCA-AAAUCUUUCAUUCGGCUUCGACUUUGUUGCCGAUACUAUAUACAUAGUCACCUAUAGUCU ....((((.((((((........((((((...---.))))))....-...........(((((..((....))..)))))............))))))....)))). ( -19.10, z-score = -1.00, R) >droSim1.chr3R 22107884 103 - 27517382 CCUCGAUUUGUGAUUCUUAAUUUGCUUUUUGU---GGAAAGCGUCA-AAAUCUUUCAUUCGGCUUCGACUUUGUUGCCGAUACUAUAUACAUAGUCGCCUAUAGUCU ....(((((.((((.(((..((..(.....).---.))))).))))-)))))......(((((..((....))..))))).((((((..(......)..)))))).. ( -19.10, z-score = -0.66, R) >droSec1.super_13 1060297 103 - 2104621 CCUCGAUUUGUGAUUCUUAAUUUGCUUUUUGU---GGAAAGCGUCA-AAAUCUUUCAUUCGGCUUCGACUUUGUUGCCGAUACUAUAUACAUAGUCGCCUAUAGUCU ....(((((.((((.(((..((..(.....).---.))))).))))-)))))......(((((..((....))..))))).((((((..(......)..)))))).. ( -19.10, z-score = -0.66, R) >droYak2.chr3R 4612338 95 + 28832112 CCUCGAUUUGUGAUUCUUAAUUUGCUUUUUGU---GGAAAGCGUCA-AAAUCUUUCAAUCGGCUUCGACUUUGUUGCCGAUACUAUAUGCUAUAUAGUC-------- ....(((((.((((.(((..((..(.....).---.))))).))))-))))).....((((((..((....))..))))))(((((((...))))))).-------- ( -20.80, z-score = -2.04, R) >droWil1.scaffold_181130 7128788 99 - 16660200 UUUCGAUUUGUGAUUUUUAAUUUGCUUUUUGUGUGGAAAAGCGUCAAAAAAUUGUUCAUUUG-AACGAUUUUUUUCUUGUUGUAUCGCUUGGUGUUGUUU------- ...((((..(((((...((((..(((((((.....)))))))...((((((((((((....)-)))))))))))....)))).))))).....))))...------- ( -20.80, z-score = -2.17, R) >droVir3.scaffold_13047 4810320 83 - 19223366 UUUCGAUUUCUGAUUCCUUAUUUGCUUUUUGU---GGAAAGCAUCAGAAAUCUGCUGCUUCUUCUUGGACUUAAUGCGACUGUCGU--------------------- ....((((((((((..(((.((..(.....).---.))))).))))))))))...............(((...........)))..--------------------- ( -17.60, z-score = -1.28, R) >consensus CCUCGAUUUGUGAUUCUUAAUUUGCUUUUUGU___GGAAAGCGUCA_AAAUCUUUCAUUCGGCUUCGACUUUGUUGCCGAUACUAUAUACAUAGUCGCCU_______ ....(((((.((((.........(((((((.....))))))))))).)))))......(((((............)))))........................... (-11.38 = -11.17 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:39 2011