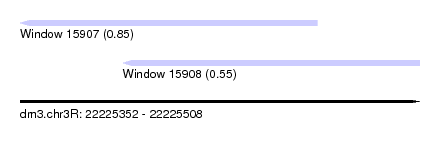
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,225,352 – 22,225,508 |
| Length | 156 |
| Max. P | 0.846820 |

| Location | 22,225,352 – 22,225,468 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.63 |
| Shannon entropy | 0.53915 |
| G+C content | 0.52956 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -12.03 |
| Energy contribution | -13.00 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.846820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22225352 116 - 27905053 GCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCUGACAGCCACACUCACACUAUGACGCACACACACACACACUAGAAUACAC ...((((((((((((.(((((((((..........))).))))))))))).............(((.....))).............))).))))..................... ( -31.50, z-score = -2.47, R) >droSim1.chr3R 22063046 116 - 27517382 GCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCUGACAGCCACACUCACACUAUGACACACACACACACACACUAGAAUACAC ...((((((((((((.(((((((((..........))).))))))))))).............(((.....))).............))).))))..................... ( -31.50, z-score = -2.94, R) >droSec1.super_13 1011382 114 - 2104621 GCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCUGACAGCCACACUCACACUAUG--ACACACACACACACACUAGAAUACAC ...((((((((((((.(((((((((..........))).))))))))))).............(((.....))).............)--)).))))................... ( -31.50, z-score = -2.85, R) >droYak2.chr3R 4567953 116 + 28832112 GCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCUGACAGCCACACUCACACUAUGAGACACACUCACUCACACCAGAACACAC ((..(((((.(((((.(((((((((..........))).)))))))))))...)))))..)).(((.....)))...((((.....)))).......................... ( -31.40, z-score = -2.24, R) >droEre2.scaffold_4820 4643598 112 + 10470090 GCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCUGACAGCCGCACUCACACUAUGACACACA----CUCACACCACAACACAC ...((((((((((((.(((((((((..........))).)))))))))))..........((.(((.....)))))...........))).))))----................. ( -33.50, z-score = -3.05, R) >droAna3.scaffold_13088 98530 97 - 569066 GCAUGUGGUCGGUUGACCGGGCCA--AAACCAGAAUUG---CCGGGAACUUAAAUCAUUCGCCGGCUGACAGCCACACACA-GCACUGAUAUACGGAGGCCAG------------- ((.((((((.(((((...((....--...))......(---(((((((.........))).)))))...))))))).))))-)).(((.....))).......------------- ( -27.80, z-score = -0.34, R) >dp4.chr2 12016049 100 + 30794189 GCAUGUGGUCGCCUGACCAGGCCA--UCACAGGAAUU---UCCACGAUCUUAAAUCAUUC--CGCCCAGCGCUGACGGACACUCCAU---ACAAGUCUGCGAGUAUAUAG------ ((.((((((.((((....)))).)--)))))(((((.---.....(((.....)))))))--).....))(((..(((((.......---....)))))..)))......------ ( -25.30, z-score = -1.00, R) >droPer1.super_3 5865509 100 + 7375914 GCAUGUGGUCGCCUGACCAGGCCA--UCACAGGAAUU---UCCACGAUCUUAAAUCAUUC--CGCCCAGCGCUGACGGACACUCCAU---ACAAGUCUGCGAGUAUAUAG------ ((.((((((.((((....)))).)--)))))(((((.---.....(((.....)))))))--).....))(((..(((((.......---....)))))..)))......------ ( -25.30, z-score = -1.00, R) >consensus GCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCUGACAGCCACACUCACACUAUGA_ACACACACACACACACUAGAA_ACAC ((.((((((((((((((((((...................))))).........)))...))))))).)))))........................................... (-12.03 = -13.00 + 0.97)

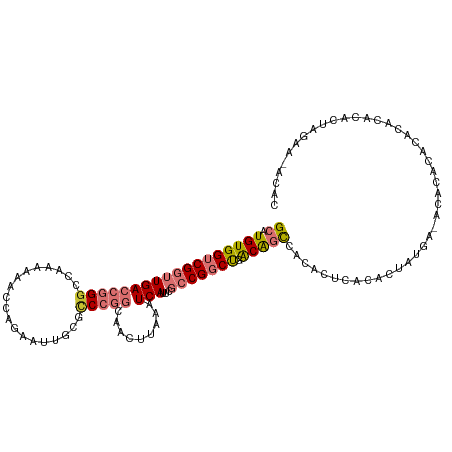
| Location | 22,225,392 – 22,225,508 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.16 |
| Shannon entropy | 0.26131 |
| G+C content | 0.53833 |
| Mean single sequence MFE | -40.86 |
| Consensus MFE | -25.79 |
| Energy contribution | -26.51 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22225392 116 - 27905053 UUUCCCGUGAUUUAGCCGGACAGUUUGAAAUAUGCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUU-CGCC---GGCUGACAGCCAC ......(((((((((((((.((((.........)))))))))........(((((.(((((((((..........))).))))))))))))))))))).-....---(((.....))).. ( -44.50, z-score = -2.87, R) >droSim1.chr3R 22063086 116 - 27517382 UUUCCCGUGAUUUAGCCGGACAGUUUGAAAUAUGCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUU-CGCC---GGCUGACAGCCAC ......(((((((((((((.((((.........)))))))))........(((((.(((((((((..........))).))))))))))))))))))).-....---(((.....))).. ( -44.50, z-score = -2.87, R) >droSec1.super_13 1011420 116 - 2104621 UUUCCCAUGAUUUAGCCGGACAGUUUGAAAUAUGCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUU-CGCC---GGCUGACAGCCAC ......(((((((((((((.((((.........)))))))))........(((((.(((((((((..........))).))))))))))))))))))).-....---(((.....))).. ( -44.20, z-score = -3.04, R) >droYak2.chr3R 4567993 116 + 28832112 UUUCCCGUGAUUUAGCCGGACAGUUUGAAAUAUGCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUU-CGCC---GGCUGACAGCCAC ......(((((((((((((.((((.........)))))))))........(((((.(((((((((..........))).))))))))))))))))))).-....---(((.....))).. ( -44.50, z-score = -2.87, R) >droEre2.scaffold_4820 4643634 111 + 10470090 -----CGUGAUUUAGCCGGACAGUUUGAAAUAUGCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUU-CGCC---GGCUGACAGCCGC -----.(((((((((((((.((((.........)))))))))........(((((.(((((((((..........))).))))))))))))))))))).-...(---(((.....)))). ( -45.60, z-score = -3.06, R) >droAna3.scaffold_13088 98556 111 - 569066 UCGCCUGUGAUUUGGCCGGACAGUUUGAAAUAUGCUGCCGGCAUGUGGUCGGUUGACCGGGCCA--AAACCAGAAUUG---CCGGGAACUUAAAUCAUU-CGCC---GGCUGACAGCCAC ..(.((((.((((((((((.((((.........)))))))))...(((((((....)).)))))--....))))...(---(((((((.........))-).))---)))).)))).).. ( -38.60, z-score = -0.95, R) >dp4.chr2 12016074 115 + 30794189 UUGGCCGUGAUUUGGCUCGACACUAUGAAAUAUGCUGCCGGCAUGUGGUCGCCUGACCAGGCCA--UCACAGGAAUU---UCCACGAUCUUAAAUCAUUCCGCCCAGCGCUGACGGACAC .((.((((....(((((((......))).....((((..(((.((((((.((((....)))).)--)))))(((((.---.....(((.....))))))))))))))))))))))).)). ( -32.50, z-score = -0.17, R) >droPer1.super_3 5865534 115 + 7375914 UUGGCCGUGAUUUGGCUCGACACUAUGAAAUAUGCUGCCGGCAUGUGGUCGCCUGACCAGGCCA--UCACAGGAAUU---UCCACGAUCUUAAAUCAUUCCGCCCAGCGCUGACGGACAC .((.((((....(((((((......))).....((((..(((.((((((.((((....)))).)--)))))(((((.---.....(((.....))))))))))))))))))))))).)). ( -32.50, z-score = -0.17, R) >consensus UUUCCCGUGAUUUAGCCGGACAGUUUGAAAUAUGCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUU_CGCC___GGCUGACAGCCAC ......(((((((((((((.((((.........)))))))))...(((((((....)).)))))..........................)))))))).........(((.....))).. (-25.79 = -26.51 + 0.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:34 2011