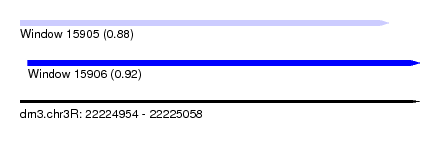
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,224,954 – 22,225,058 |
| Length | 104 |
| Max. P | 0.922124 |

| Location | 22,224,954 – 22,225,050 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.14 |
| Shannon entropy | 0.46802 |
| G+C content | 0.41752 |
| Mean single sequence MFE | -22.09 |
| Consensus MFE | -11.06 |
| Energy contribution | -11.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
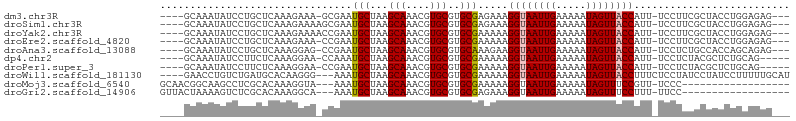
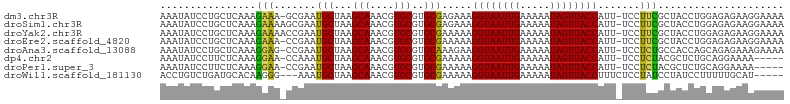
>dm3.chr3R 22224954 96 + 27905053 ----GCAAAUAUCCUGCUCAAAGAAA-GCGAAUGCUAAGCAAACGUGCGUGCGAGAAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUUCGCUACCUGGAGAG--- ----............(((..((..(-((((((((...(((....)))..))).((((((((((((.....)))))))).))-)).))))))..)).)))..--- ( -23.90, z-score = -1.85, R) >droSim1.chr3R 22062646 97 + 27517382 ----GCAAAUAUCCUGCUCAAAGAAAAGCGAAUGCUAAGCAAACGUGCGUGCGAGAAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUUCGCUACCUGGAGAG--- ----............(((.......(((((((((...(((....)))..))).((((((((((((.....)))))))).))-)).)))))).....)))..--- ( -22.80, z-score = -1.49, R) >droYak2.chr3R 4567537 97 - 28832112 ----GCAAAUAUCCUGCUCAAAGAAAACCGAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUUCGCUACCUGGAGAG--- ----(((.......)))..........(((........(((....)))((((((((((((((((((.....)))))))).))-)..))))).)).)))....--- ( -17.40, z-score = -0.41, R) >droEre2.scaffold_4820 4643198 96 - 10470090 ----GCAAAUAUCCUGCUCAAAGAAA-CCGAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUUCGCUACCUGGAGAG--- ----(((.......))).........-(((........(((....)))((((((((((((((((((.....)))))))).))-)..))))).)).)))....--- ( -17.40, z-score = -0.40, R) >droAna3.scaffold_13088 98107 96 + 569066 ----GCAAAUAUCCUGCUCAAAGGAG-CCGAAUGCUAAGCAAACGUGCGUGCAAAGAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUCUGCCACCAGCAGAG--- ----(((....((((......))))(-(((..(((...)))..)).)).)))......((((((((.....))))))))...-..((((((.....))))))--- ( -27.00, z-score = -2.88, R) >dp4.chr2 12015611 94 - 30794189 ----GCAAAUAUCCUUCUCAAAGGAA-CCAAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUCUACGCUCUGCAG----- ----(((....(((((....))))).-.....)))...(((...(.(((((.(((((.((((((((.....)))))))).))-)..))))))).))))..----- ( -20.60, z-score = -2.25, R) >droPer1.super_3 5865071 94 - 7375914 ----GCAAAUAUCCUUCUCAAAGGAA-CCGAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUCUACGCUCUGCAG----- ----(((....(((((....))))).-.....)))...(((...(.(((((.(((((.((((((((.....)))))))).))-)..))))))).))))..----- ( -20.60, z-score = -1.88, R) >droWil1.scaffold_181130 7073715 98 + 16660200 ----GAACCUGUCUGAUGCACAAGGG---AAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUACCUUUCUCCUAUCCUAUCCUUUUUGCAU ----...........(((((.(((((---(...((...(((....)))..))((.(((((((((((.....))))))))))).))........)))))).))))) ( -27.60, z-score = -3.64, R) >droMoj3.scaffold_6540 12498811 83 - 34148556 GCAACGGCAAGCCUCGCACAAAGGUA---AAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUUCCGUU-UCCC------------------ ..(((((.((((.((((((...(((.---....)))..(((....)))))))))........(((....))).)))))))))-....------------------ ( -19.90, z-score = -1.02, R) >droGri2.scaffold_14906 10191944 83 + 14172833 GUUACUAAAAGUCUCGCACAAAGGCA---AAAUGCUAAGCAAACGUGCGUGCGAGAAAGGUAAUUGAAAAAUAGUUUCCUUU-UUCC------------------ ((((((.....((((((((...(((.---....)))..(((....)))))))))))..)))))).((((((........)))-))).------------------ ( -23.70, z-score = -3.50, R) >consensus ____GCAAAUAUCCUGCUCAAAGGAA_CCGAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUACCAUU_UCCUUCGCUACCUGGAGAG___ ................................(((...(((....)))..))).....((((((((.....)))))))).......................... (-11.06 = -11.17 + 0.11)
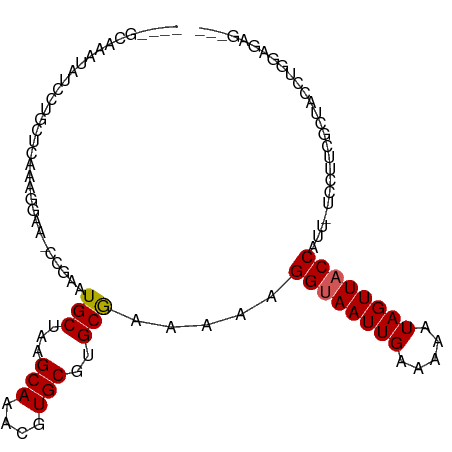
| Location | 22,224,956 – 22,225,058 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.15 |
| Shannon entropy | 0.33360 |
| G+C content | 0.39516 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -13.01 |
| Energy contribution | -12.19 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
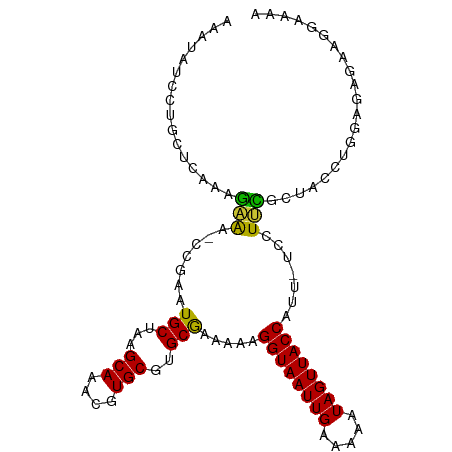
>dm3.chr3R 22224956 102 + 27905053 AAAUAUCCUGCUCAAAGAAA-GCGAAUGCUAAGCAAACGUGCGUGCGAGAAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUUCGCUACCUGGAGAGAAGGAAAA .....((((.(((..((..(-((((((((...(((....)))..))).((((((((((((.....)))))))).))-)).))))))..))...))).))))... ( -30.50, z-score = -3.76, R) >droSim1.chr3R 22062648 103 + 27517382 AAAUAUCCUGCUCAAAGAAAAGCGAAUGCUAAGCAAACGUGCGUGCGAGAAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUUCGCUACCUGGAGAGAAGGAAAA .....((((.(((..((...(((((((((...(((....)))..))).((((((((((((.....)))))))).))-)).))))))..))...))).))))... ( -29.30, z-score = -3.46, R) >droYak2.chr3R 4567539 103 - 28832112 AAAUAUCCUGCUCAAAGAAAACCGAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUUCGCUACCUGGAGAGAAGGAAAA .....((((.(((...................(((....)))((((((((((((((((((.....)))))))).))-)..))))).)).....))).))))... ( -23.10, z-score = -2.03, R) >droEre2.scaffold_4820 4643200 102 - 10470090 AAAUAUCCUGCUCAAAGAAA-CCGAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUUCGCUACCUGGAGAGAAGGAAAA .....((((.(((.......-...........(((....)))((((((((((((((((((.....)))))))).))-)..))))).)).....))).))))... ( -23.10, z-score = -1.97, R) >droAna3.scaffold_13088 98109 102 + 569066 AAAUAUCCUGCUCAAAGGAG-CCGAAUGCUAAGCAAACGUGCGUGCAAAGAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUCUGCCACCAGCAGAGAAAGAAAA .......(.((((....)))-).)..(((...(((....)))..))).....((((((((.....)))))))).((-((((((((.....))))))...)))). ( -26.40, z-score = -2.88, R) >dp4.chr2 12015613 97 - 30794189 AAAUAUCCUUCUCAAAGGAA-CCAAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUCUACGCUCUGCAGGAAAA----- .....(((((....))))).-((...(((..(((...(((....))).....((((((((.....))))))))...-........)))..)))))....----- ( -21.20, z-score = -2.05, R) >droPer1.super_3 5865073 97 - 7375914 AAAUAUCCUUCUCAAAGGAA-CCGAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUACCAUU-UCCUCUACGCUCUGCAGGAAAA----- .....(((((....))))).-((...(((..(((...(((....))).....((((((((.....))))))))...-........)))..)))))....----- ( -21.20, z-score = -1.74, R) >droWil1.scaffold_181130 7073717 96 + 16660200 ACCUGUCUGAUGCACAAGGG---AAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUACCUUUCUCCUAUCCUAUCCUUUUUGCAU----- .........(((((.(((((---(...((...(((....)))..))((.(((((((((((.....))))))))))).))........)))))).)))))----- ( -27.60, z-score = -3.76, R) >consensus AAAUAUCCUGCUCAAAGAAA_CCGAAUGCUAAGCAAACGUGCGUGCGAAAAAGGUAAUUGAAAAAUAGUUACCAUU_UCCUUCGCUACCUGGAGAGAAGGAAAA ................(((.......(((...(((....)))..))).....((((((((.....)))))))).......)))..................... (-13.01 = -12.19 + -0.83)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:33 2011