
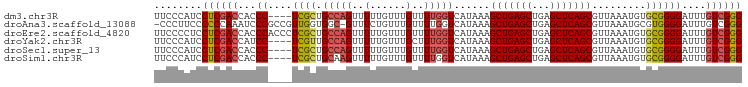
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,195,688 – 22,195,782 |
| Length | 94 |
| Max. P | 0.838765 |

| Location | 22,195,688 – 22,195,782 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 90.18 |
| Shannon entropy | 0.18922 |
| G+C content | 0.51559 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -24.17 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22195688 94 + 27905053 UUCCCAUCCUCGACCACCC----UCGCUGCCAGUUUUUGUUUGUUUUGGUCAUAAAGCUGAGCUGAGCUCAGCGUUAAAUGUGCGGGGAUUUGUCGGG ........((((((...((----((((.(((((..(......)..)))))......(((((((...))))))).........))))))....)))))) ( -32.30, z-score = -2.38, R) >droAna3.scaffold_13088 78093 96 + 569066 -CCCUUCCCCCCAAAUCCCGCCGUUGGUGGC-GUUUCUGUUUGUUUUGGUCAUAAAGCUGAGCUGAGCUCAGCGUUAAAUGCGUGGGGAUUUGUCGGG -.....(((..((((((((..(((...((((-.......(((((.......)))))(((((((...)))))))))))...)))..))))))))..))) ( -35.20, z-score = -2.59, R) >droEre2.scaffold_4820 4623181 98 - 10470090 UUCCCCUCCUCGACCACCCACCCUCGCUGCCAGUUUUUGUUUGUUUUGGUCAUAAAGCUGAGCUGAGCUCAGCGUUAAAUGUGCGGGGAUUUGUCGGG ........((((((.......((((((.(((((..(......)..)))))......(((((((...))))))).........))))))....)))))) ( -30.90, z-score = -1.58, R) >droYak2.chr3R 4546556 94 - 28832112 UUCCCAUCCUCGACCAUCC----UCGUUGCCAGUUUUUGUUUGCUUUGGUCAUAAAGCUGAGCUGAGCUCAGCGUUAAAUGUGCGGGGAUUUGUCGGG ........((((((.((((----((((.(((((............)))))......(((((((...))))))).........))))))))..)))))) ( -32.80, z-score = -2.73, R) >droSec1.super_13 983389 94 + 2104621 UUCCCAUCCUCGACCACCC----UCGCUGCCAGUUUUUGUUUGUUUUGGUCAUAAAGCUGAGCUGAGCUCAGCGUUAAAUGUGCGGGGAUUUGUCGGG ........((((((...((----((((.(((((..(......)..)))))......(((((((...))))))).........))))))....)))))) ( -32.30, z-score = -2.38, R) >droSim1.chr3R 22042433 94 + 27517382 UUCCCAUCCUCGACCACCC----UCGCUGCAAGUUUUUGUUUGUUUUGGUCAUAAAGCUGAGCUGAGCUCAGCGUUAAAUGUGCGGGGAUUUGUCGGG ........((((((...((----((((.(((((......)))))............(((((((...))))))).........))))))....)))))) ( -30.10, z-score = -1.74, R) >consensus UUCCCAUCCUCGACCACCC____UCGCUGCCAGUUUUUGUUUGUUUUGGUCAUAAAGCUGAGCUGAGCUCAGCGUUAAAUGUGCGGGGAUUUGUCGGG ..((((((((((..(((.......(((((..(((((..((((((((((....)))))).)))).))))))))))......)))))))))).....))) (-24.17 = -24.70 + 0.53)

| Location | 22,195,688 – 22,195,782 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 90.18 |
| Shannon entropy | 0.18922 |
| G+C content | 0.51559 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -19.59 |
| Energy contribution | -20.59 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22195688 94 - 27905053 CCCGACAAAUCCCCGCACAUUUAACGCUGAGCUCAGCUCAGCUUUAUGACCAAAACAAACAAAAACUGGCAGCGA----GGGUGGUCGAGGAUGGGAA ((((((....((((((.........(((((((...)))))))....((.(((..............)))))))).----)))..)))).))....... ( -31.54, z-score = -2.84, R) >droAna3.scaffold_13088 78093 96 - 569066 CCCGACAAAUCCCCACGCAUUUAACGCUGAGCUCAGCUCAGCUUUAUGACCAAAACAAACAGAAAC-GCCACCAACGGCGGGAUUUGGGGGGAAGGG- (((..((((((((............(((((((...)))))))........................-(((......)))))))))))..))).....- ( -35.20, z-score = -3.93, R) >droEre2.scaffold_4820 4623181 98 + 10470090 CCCGACAAAUCCCCGCACAUUUAACGCUGAGCUCAGCUCAGCUUUAUGACCAAAACAAACAAAAACUGGCAGCGAGGGUGGGUGGUCGAGGAGGGGAA ((((((....((((((.........(((((((...)))))))....((.(((..............)))))))).)))......)))).))....... ( -31.04, z-score = -1.28, R) >droYak2.chr3R 4546556 94 + 28832112 CCCGACAAAUCCCCGCACAUUUAACGCUGAGCUCAGCUCAGCUUUAUGACCAAAGCAAACAAAAACUGGCAACGA----GGAUGGUCGAGGAUGGGAA ((((((..((((.((..........(((((((...)))))))....((.(((..............))))).)).----)))).)))).))....... ( -26.24, z-score = -1.73, R) >droSec1.super_13 983389 94 - 2104621 CCCGACAAAUCCCCGCACAUUUAACGCUGAGCUCAGCUCAGCUUUAUGACCAAAACAAACAAAAACUGGCAGCGA----GGGUGGUCGAGGAUGGGAA ((((((....((((((.........(((((((...)))))))....((.(((..............)))))))).----)))..)))).))....... ( -31.54, z-score = -2.84, R) >droSim1.chr3R 22042433 94 - 27517382 CCCGACAAAUCCCCGCACAUUUAACGCUGAGCUCAGCUCAGCUUUAUGACCAAAACAAACAAAAACUUGCAGCGA----GGGUGGUCGAGGAUGGGAA ((((((....((((((.........(((((((...))))))).................(((....)))..))).----)))..)))).))....... ( -29.00, z-score = -2.29, R) >consensus CCCGACAAAUCCCCGCACAUUUAACGCUGAGCUCAGCUCAGCUUUAUGACCAAAACAAACAAAAACUGGCAGCGA____GGGUGGUCGAGGAUGGGAA ((((((....(((............(((((((...)))))))....((.(((..............)))))........)))..)))).))....... (-19.59 = -20.59 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:25 2011